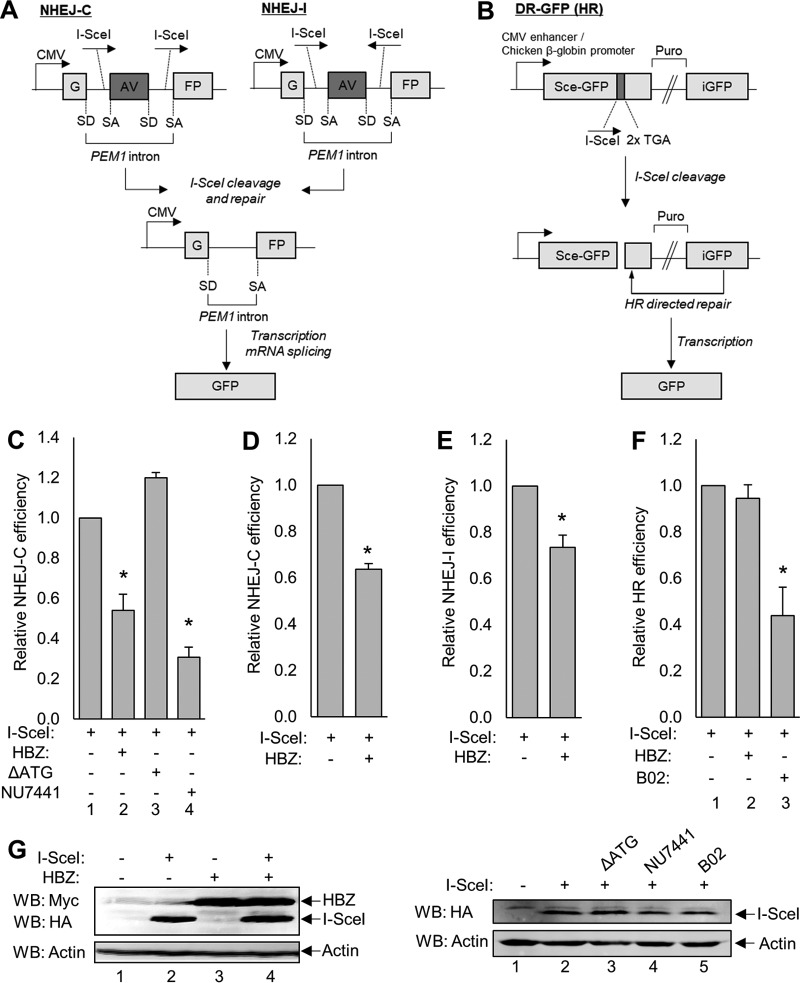
FIG 2.
GFP reporter-based DSB repair vectors reveal that HBZ attenuates NHEJ-C and NHEJ-I but not HR. (A) Schematics of the reporter vectors used to measure NHEJ of compatible DSBs (NHEJ-C) and NHEJ of incompatible breaks (NHEJ-I), both of which are part of the classical NHEJ pathway. The GFP reporter gene is separated by the PEM1 gene intron containing an adenoviral (AV) exon flanked by two I-SceI restriction sites in a direct orientation (NHEJ-C) or an indirect orientation (NHEJ-I). Cleavage at these sites results in the excision of the AV exon and repair by NHEJ. The PEM1 intron is spliced out of the transcript at the splice donor (SD) site and splice acceptor (SA) site, resulting in an uninterrupted GFP-encoding transcript. CMV, cytomegalovirus. (B) Schematic of the reporter vector used to measure HR. This vector contains one complete copy of the GFP reporter gene (Sce-GFP) and a truncated copy of GFP that serves as a template for repair (iGFP), which are separated by a puromycin resistance cassette (Puro). Eleven base pairs were deleted from the Sce-GFP cDNA sequence and replaced with one I-SceI restriction site and two in-frame stop codons. This missing sequence ensures that template-dependent repair must occur to result in the production of a full-length GFP gene. (C) HEK 293T cells were transfected with a total of 6 μg of plasmid DNA (2 μg NHEJ-C reporter, 2 μg pSG-I-SceI-HA, 2 μg pcDNA-HBZ-Myc-His or pcDNA-ΔATG-Myc-His, or 2 μg empty vector) and treated with 2 μM NU7441 where indicated. Cells were collected for flow cytometric analysis of GFP expression, as outlined in Materials and Methods. Data were collected from 20,000 individual events and are shown as fold changes from the maximum repair activity (I-SceI plus reporter). HBZ data are averages of results from 10 independent experiments, ΔATG data are averages of results from three independent experiments, and NU7441 data are averages of results from 3 independent experiments. Error bars represent SEM (*, P ≤ 0.05 by Student's t test). (D) Jurkat cells were electroporated with a total of 15 μg of plasmid DNA (5 μg pcDNA-HBZ-Myc-His or empty vector and 10 μg NHEJ-C I-SceI-digested reporter) and analyzed for GFP expression. Data shown are averages of results from two independent experiments, and error bars indicate SEM (*, P ≤ 0.05 by Student's t test). (E) HEK 293T cells were transfected with a total of 6 μg of plasmid DNA (2 μg pSG-I-SceI-HA, 2 μg pcDNA-HBZ-Myc-His or empty vector, and 2 μg NHEJ-I reporter) and analyzed as outlined above. Data shown are averages of results from five independent experiments, and error bars represent SEM (*, P ≤ 0.05 by Student's t test). (F) HEK 293T cells were transfected with a total of 6 μg of plasmid DNA (2 μg pSG-I-SceI-HA, 2 μg pcDNA-HBZ-Myc-His or empty vector, and 2 μg the pDR-GFP reporter) and treated with 20 μM B02 for 48 h where indicated. Data shown are averages of results from three independent experiments, and error bars represent SEM (*, P ≤ 0.05 by Student's t test). (G) Transfected cellular protein extracts (30 to 60 μg) were analyzed by Western blotting for HBZ and/or I-SceI expression using the indicated antibodies. HA, hemagglutinin.