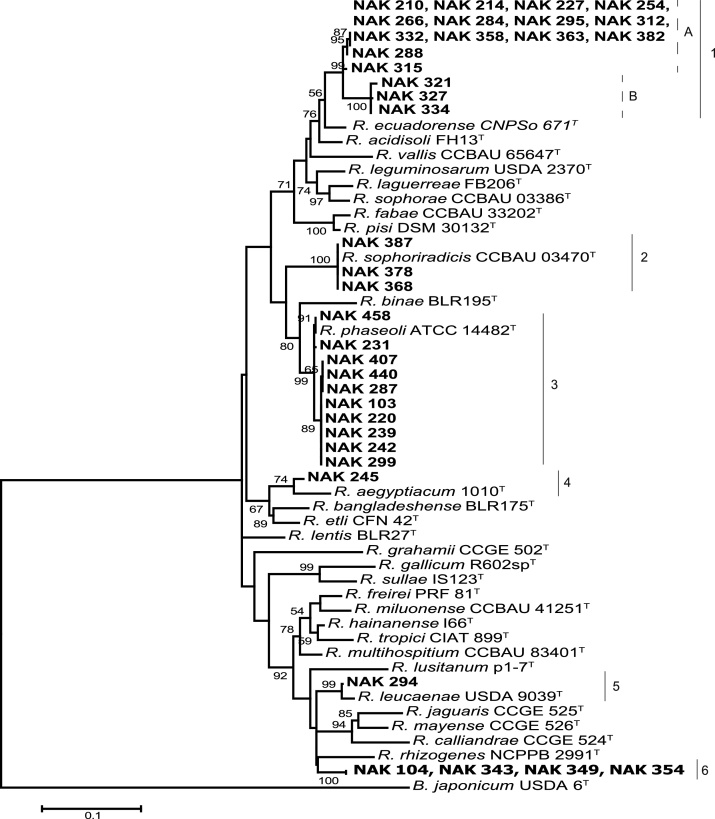
Fig. 2.
The phylogenetic relationship between the study isolates (in bold) and type strains of closely related species based on concatenated recA and atpD genes. The evolutionary history was inferred using the Maximum Likelihood method based on the General Time Reversible model in MEGA6 [53]. There was a total of 736 positions in the final dataset, and node supports higher than 50% are labelled with a bootstrap value (1000 replicates). The sequence of B. japonicum USDA 6T was included as an outgroup. Bar indicates 10 nucleotide substitutions per 100 nucleotides.