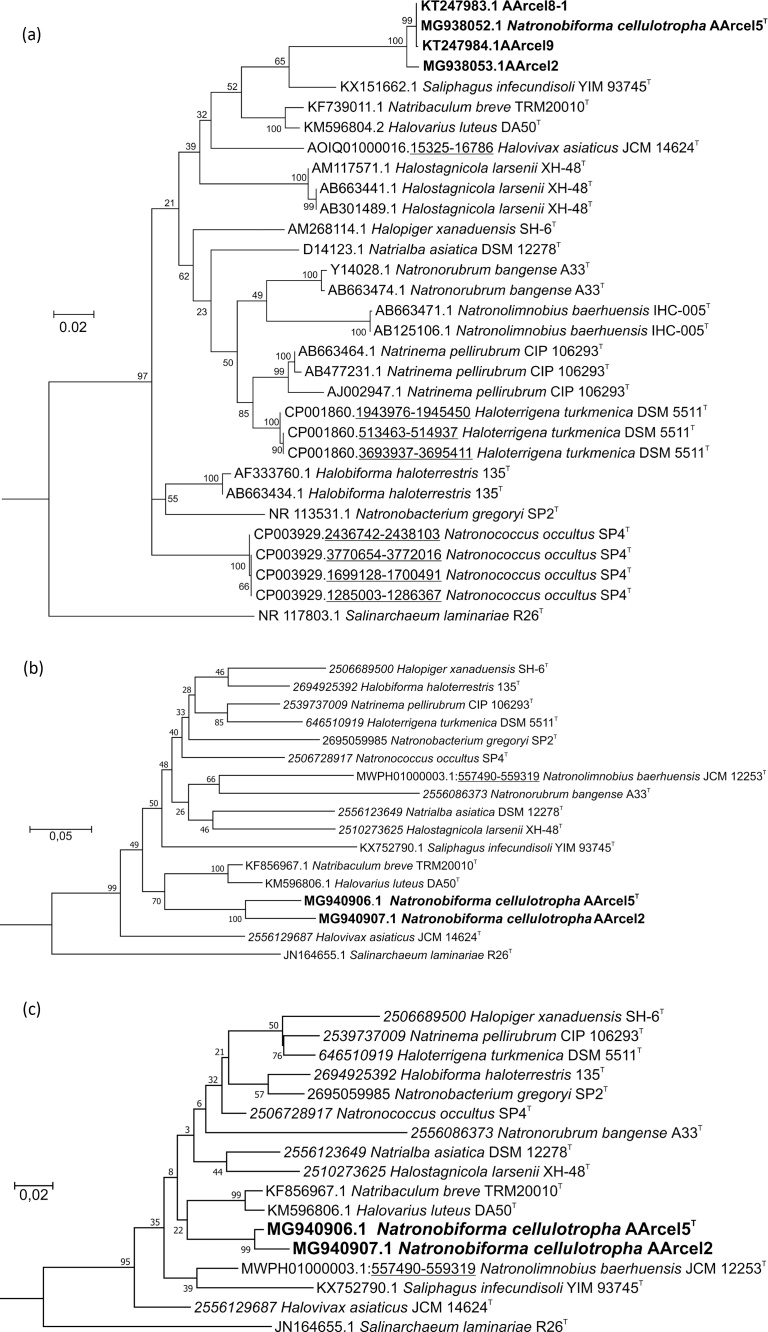
Fig. 3.
Phylogeny of the AArcel strains. (a) Maximum Likelihood 16S rRNA gene sequence-based phylogenetic tree showing position of the AArcel strains (in bold) within the family Natrialbaceae. Branch lengths (see scale) correspond to the number of substitutions per site with corrections, associated with the model (GTR, G + I, 4 categories). All positions with less than 95% site coverage were eliminated. Totally 1359 positions were used in the alignment of 32 sequences (except for the partial AArcel4 and AArcel6 sequences, 100% identical to AArcel5T). Numbers at nodes indicate bootstrap values of 1000 repetitions. Halomarina oriensis strain JCM 16495 (AB663390.1) was used as an outgroup. (b) Maximum Likelihood rpoB' gene sequence-based tree showing position of the AArcel2 and AArcel5 strains (in bold) within family Natrialbaceae. Totally 1827 positions were used in the alignment of 18 sequences. Halomarina oriensis JCM 16495 (KJ870934.1) was used as an outgroup. (c): Maximum Likelihood RpoB' protein sequence-based tree showing position AArcel2 and AArcel5 strains (in bold) within the family Natrialbaceae. Totally 608 positions were used in the alignment of 18 sequences. Halomarina oriensis JCM 16495 (KJ870934.1) was used as an outgroup. Sequences with accession numbers in italic were obtained from IMG, in roman – from the Genbank.