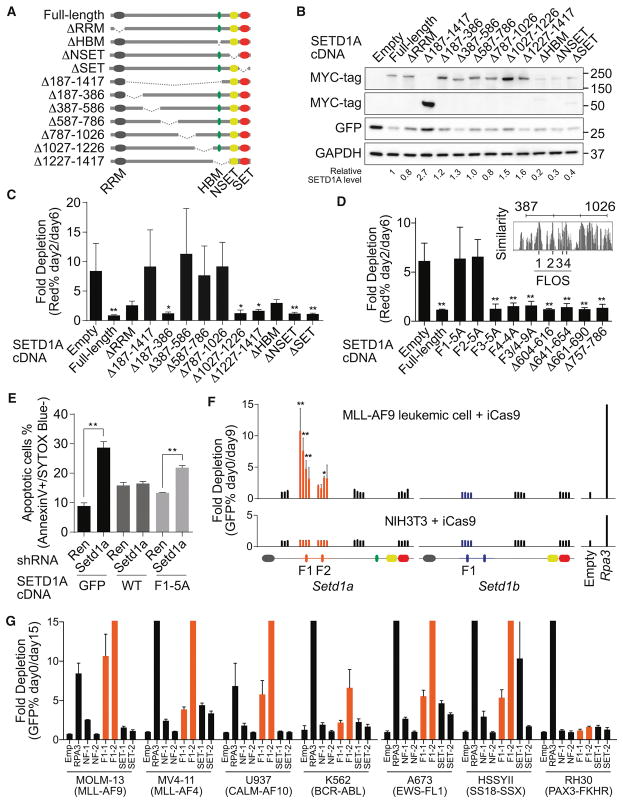
Figure 2. Internal region on SETD1A encodes a critical functional domain for leukemia cell survival.
A. The human SETD1A deletion mutant constructs are shown as schematic illustrations.
B. Western blot analysis was performed using SETD1A deletion mutant-expressing 293T cells. These constructs were used for the following rescue experiments. Representative images from 2 independent experiments are shown.
C. Human SETD1A deletion mutant-expressing mouse MLL-AF9 leukemia cells were transfected with dox-inducible mouse Setd1a 3′UTR targeting shRNAs. This experiment was performed with 2 independent shRNAs and repeated 2 times with 3 biological replicates in each experiment.
D. Evolutionarily conserved motifs were identified inside of the functional region on SETD1A and named as FLOS1-4 (upper right panel). SETD1A mutant-expressing mouse MLL-AF9 leukemia cells were transfected with mouse Setd1a shRNAs. This experiment was performed with 2 independent shRNAs and repeated 2 times with 3 biological replicates in each experiment.
E. Empty (GFP), SETD1A wild-type (WT) and FLOS1/alanine mutant (F1-5A)–expressing cells were transfected with mouse Setd1a 3′UTR shRNAs, and the annexin V+DAPI− populations were analyzed at 4 days post-dox. This experiment was performed with 2 independent shRNAs and repeated 2 times with 3 biological replicates in each experiment.
F. Domain-focused CRISPR strategy was performed with Setd1a or Setd1b sgRNAs in inducible Cas9 (iCas9)-expressing MLL-AF9 leukemia cells or NIH3T3 cells. The relative location of each sgRNA to the SETD1A or SETD1B protein is indicated along the x-axis. Empty vector or Rpa3 sgRNAs were used as negative or positive controls, respectively. Representative data from one out of 3 independent experiments are shown.
G. Domain-focused CRISPR strategy was performed with Setd1a sgRNAs in human leukemia and sarcoma cell lines. Empty vector or RPA3 sgRNAs were used as negative or positive controls, respectively. This experiment was repeated 2 times with 3 biological replicates in each experiment.