Figure 1.
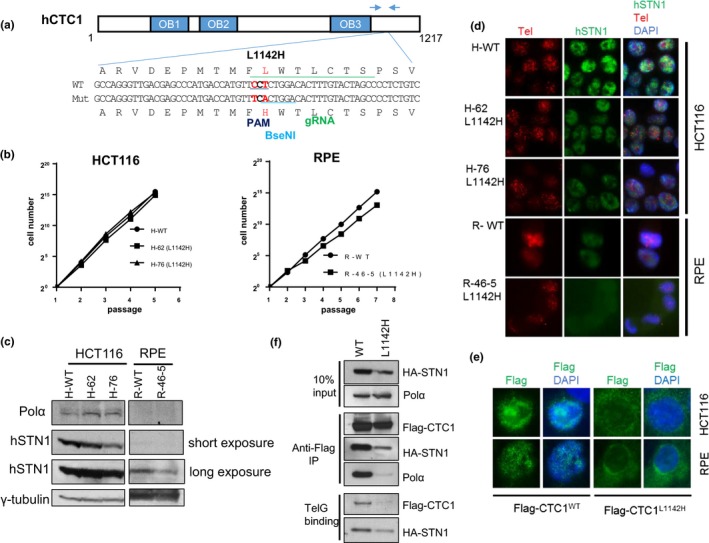
Generation of the CTC1 L1142H mutation in HCT116 and RPE cells using CRISPR/Cas9. (a) Schematic of the guide sgRNA utilized to mutate CTC1L1142 to CTC1H1142. Arrows indicate PCR primers used for genotyping. (b) NIH 3T3 assays were used to measure the proliferative capacities of the indicated cell lines. (c) Expression pattern of endogenous DNA Pol‐α and STN1 in the indicated cell lines detected by western analysis. γ‐tubulin was used as a loading control. (d) Immuno‐FISH analysis for endogenous STN1 (green) and telomeres (red) in WT or L1142H mutant HCT116 or RPE cell lines. STN1 was visualized using an anti‐STN1 antibody, telomeres visualized by hybridization with a 5′‐Tam‐OO‐(CCCTAA)4‐3′ PNA probe, and nuclei visualized by 4,6‐diamidino‐2‐phenylindole staining (DAPI; blue). (e) Immunostaining for WT Flag‐CTC1 or Flag‐CTC1L1142H (green) expressed in HCT116 or RPE cells. Nuclei were stained with DAPI (in blue). (f) Co‐IP was used to determine the ability of WT Flag‐CTC1 and Flag‐CTC1L1142H mutant proteins to interact with HA‐STN1, endogenous DNA pol‐α, and ss telomeric DNA [Tel‐G oligo: (TTAGGG)3]
