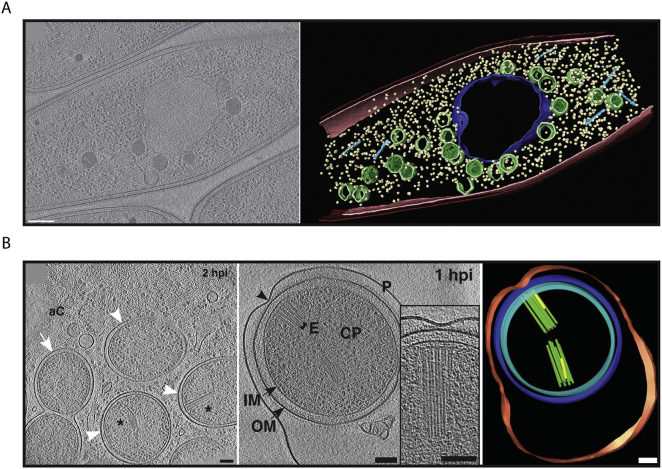
Fig. 2.
(A) Single slice through a cryo-ET reconstruction of a 201φ2–1 phage-infected P. chlororaphis prepared using FIB milling, in which filled (darker) assembled capsids are docked to an apparently contiguous shell during DNA encapsidation. A segmentation of the complete tomogram is shown (right), highlighting the shell (dark blue/purple), capsids (green), cytoplasmic membrane (pink), outer membrane (red), phage tails (light blue), and ribosomes (yellow). Scale bar; 200 nm. Adapted with permission from [42]. (B) A. asiaticus bacteria were imaged inside cells using FIB milling and cryo-ET. A representative tomographic slice is shown at later time points post infection (0.5 to 2 h post infection (hpi)) in which amoebophili are found in the amoeba cytosol (aC) (white arrowheads). Amoebophili differentiate into rods (white arrows), although some show signs of degradation (black arrowheads) and do not escape (left). At earlier time points (0.25 h post infection) amoebophili reside within phagosomes, and extended T6SS arrays can be found contacting the phagosome membrane (black arrowhead) (center), highlighted in the corresponding segmentation (right). P/red: phagosome; OM/blue: outer membrane; IM/cyan: inner membrane; CP: cytoplasm; E/green: extended T6SS; yellow: contracted T6SS. Scale bars: 100 nm. Adapted with permission from [43].