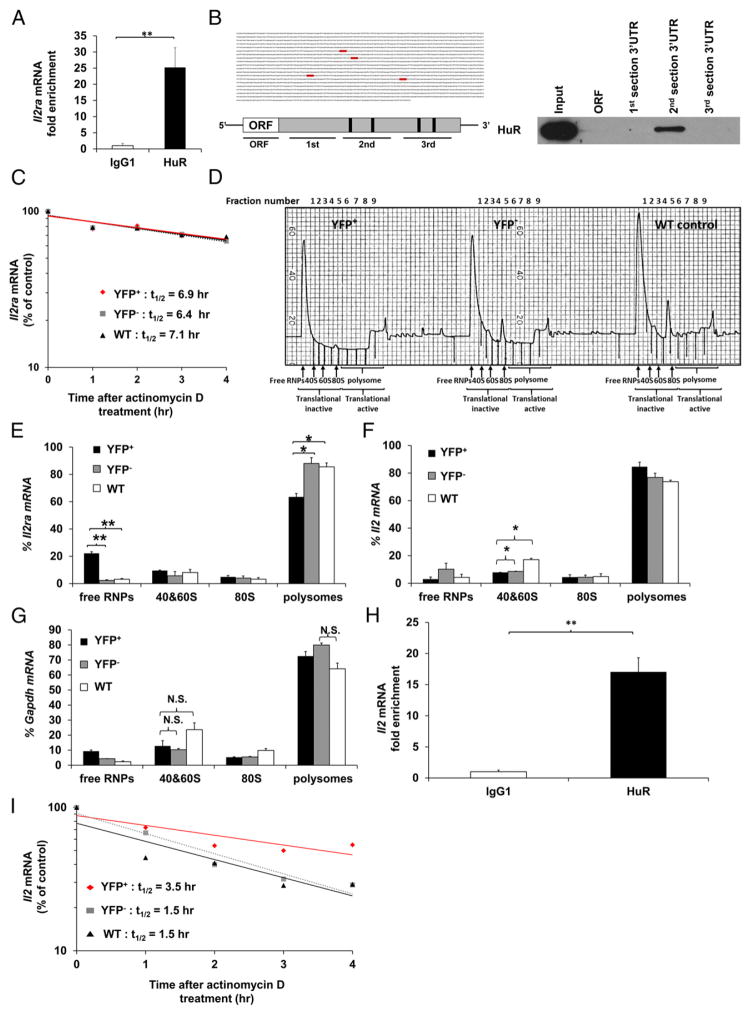
FIGURE 5. HuR physically interacts with the Il2ra 3′UTR mRNA and enhances its translational efficiency in activated CD4+ T cells.
(A) RIP using HuR or IgG1 Ab, followed by RT-PCR to determine physical HuR mRNA targets. Data are fold enrichment of Il2ra mRNA in anti-HuR samples compared with IgG1 controls. (B) Putative HuR targets, ARE elements (gray), present in the 3′UTR of Il2ra mRNA (left panels) and biotin pull-down shows association of HuR with different portions of Il2ra mRNA (right panel). Data show HuR interaction with the second section of Il2ra 3′UTR mRNA containing the first two putative HuR binding sites. (C) Il2ra mRNA stability assay in activated YFP+, YFP−, and WT CD4+ T cells on day 4 postactivation. Data represent the percentage of mRNA remaining over time after actinomycin D treatment. (D) Absorbance profile for RNA separated by velocity sedimentation through a sucrose gradient. RNA was extracted from each fraction. Polysomal gradient analysis of Il2ra (E), Il2 (F), and Gapdh (G) mRNAs in activated YFP+, YFP−, and WT CD4+ T cells. Data are the percentage of Il2ra, Il2, and Gapdh mRNA distribution in 40S, 60S, 80S, and polysome fractions by RT-PCR. (H) RIP using HuR or IgG1 Ab, followed by RT-PCR to determine HuR mRNA targets. Data are fold enrichment of Il2 mRNA in anti-HuR samples compared with IgG1 controls. (I) Il2 mRNA stability assay in activated YFP+, YFP−, and WT CD4+ T cells on day 4 postactivation. Data are from three (A and H) or two (D, F, and G) independent experiments or are a representative of three (C and I) or two (B and D) independent experiments. Data are mean + SEM of three (A and H) or two (D, F, and G) independent experiments. *p < 0.05, **p < 0.01, two-tailed unpaired t test (A and H), one-way ANOVA with the Tukey multiple-comparisons test (E–G). N.S., not significant.