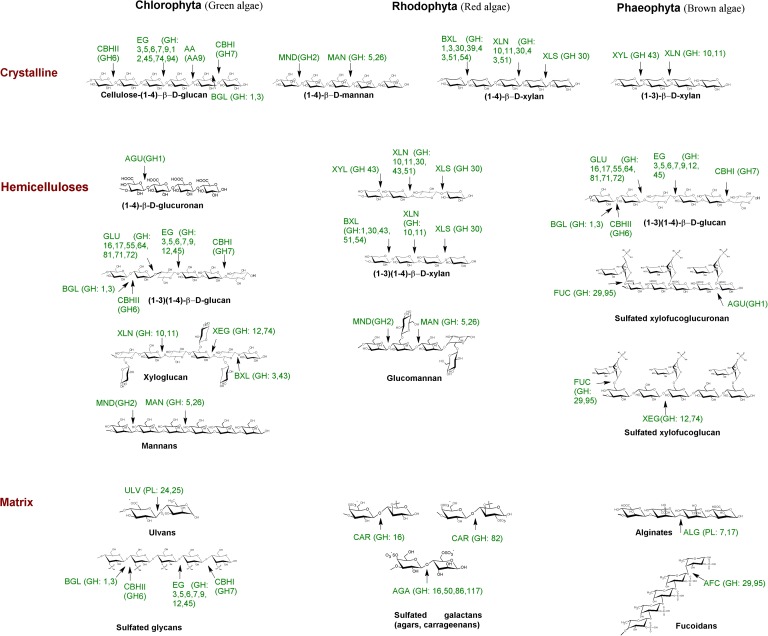
FIGURE 1.
Schematic representation of algae cell wall polysaccharides and corresponding polysaccharide-degrading enzymes. AA, monooxygenase; AFC, α-fucosidase; AGA, agarase; AGU, gucuronidase; ALG, alginate lyase; BGL, β-1,4-glucosidase; BXL, β-1,4-xylosidase; CAR, carrageenase; CBHI, exo-β-glucanase (reducing end); CBHIl, exo-β-glucanase cellobiohydrolase (non-reducing end); EG, endo-β-1,4-glucanase; GLU, β-1,4/1,3-glucanase; MAN, β-1,4-endomannanase; MND- β-1,4-mannosidase; ULV, ulvanlyase; XEG, xyloglucan-β-1,4-endoglucanase; XLN, β-1,4/1,3-endoxylanase; XLS, β-1,4-xylosidase (reducing end); XYL, β-1,3-xylosidase (van den Brink and de Vries, 2011; Pluvinage et al., 2013; Rytioja et al., 2014; Zhao et al., 2014; Zhu et al., 2016; Gao et al., 2017; Ulaganathan et al., 2017). Algae polysaccharides content and structures were presented with the use of data reported by Synytsya et al. (2015). Enzyme abbreviations were applied according the EC and CAZy classification. Algae polysaccharide-degrading enzymes were shortened according to the first letters of the enzymatic names.