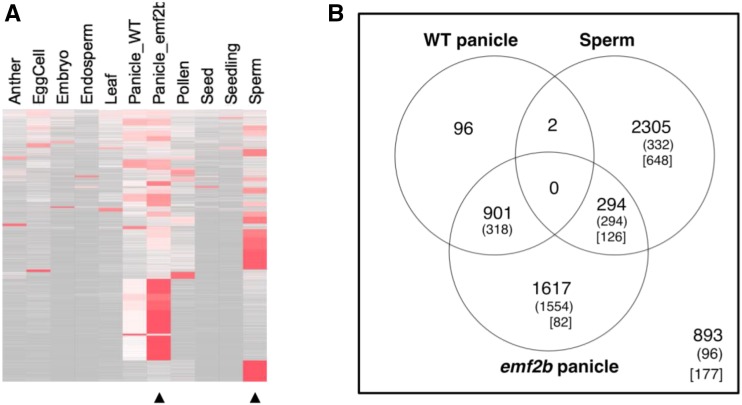
Figure 7.
A, Heat map for RPKM values row normalized to 1 across 11 tissue/cell types for the 6,213 lincRNAs (original isoform set) grouped into 39 clusters ranked top to bottom by decreasing average Shannon entropy of each cluster. Sperm-specific and emf2b panicle mutant derepressed lincRNA subclasses form a few large blocks of clusters with lower Shannon entropy (higher tissue specificity). B, Dominant expression Venn diagram for 6,108 lincRNAs with reads present within at least wild-type (WT) panicles, emf2b panicles, or sperm cells from mature pollen. LincRNAs are grouped into one of six possible sets for differentially expressed isoforms by using six different contrasts of a negative binomial model of differential expression using the edgeR package (see “Materials and Methods”). Numbers in parentheses refer to those 2,594 lincRNAs derepressed in emf2b panicles relative to wild-type panicles (as determined in the wild-type panicle versus emf2b panicle pairwise analysis) of the lincRNAs belonging to the set of 5,512 expressed in panicles. Numbers in square brackets represent the 1,033 lincRNAs with reads in emf2b panicles but not in wild-type panicles. The set of 893 refers to those not considered differentially expressed in this analysis.