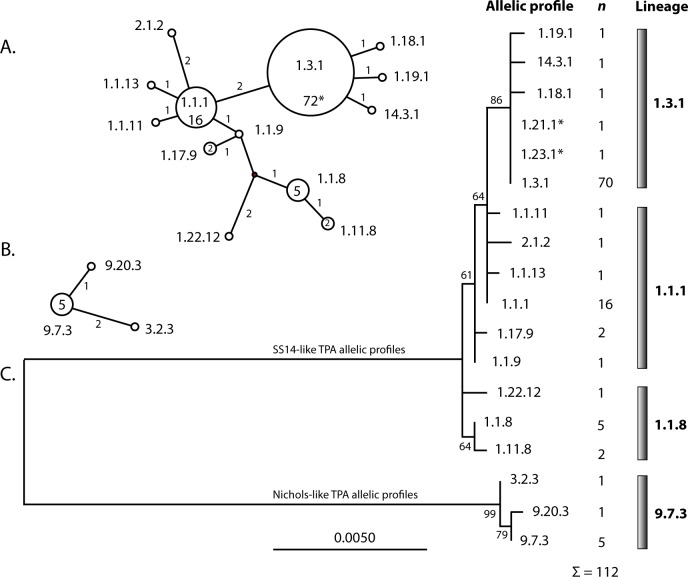
Fig 4. Phylogeny of allelic profiles of fully typed samples identified in this study.
A. The Median joining network tree of concatenated sequences of TP0136, TP0548, and TP0705 of the SS14-like clinical samples (2526 nt in length in TPA SS14). The analyzed coordinates correspond to 336–1385 of TPANIC_0136, 150–951 of TPANIC_0548, and 1453–2126 of TPANIC_0705 (TPA Nichols; CP004010.2). Every circle represents a different allelic profile and specific allelic profiles are shown within or near the circles. The number of mutations are shown next to the branch lines. The inferred haplotype is shown as a black (connecting) circle. The size of the circle represents the number of identified samples and the corresponding numbers of samples are shown inside the circles (if greater than one). B.The Median joining network tree of concatenated sequences of TP0136, TP0548, and TP0705 of the Nichols-like clinical samples. Every circle represents a different allelic profile and specific allelic profiles are shown near the circles. The number of mutations are shown next to the branch lines. A contiguous indel was considered to be a single event. The size of the circle represents the number of identified samples and the corresponding number of samples are shown inside the circles (if greater than one). C. A phylogenetic tree constructed from concatenated sequences TP0136, TP0548, and TP0705. *Sequences of samples representing allelic profiles 1.21.1 and 1.23.1 also comprise nucleotide differences in positions 96 and 127 in TP0548 that were not used for the construction of the tree, respectively. Since sequences from additional samples were not obtained in this region, both allelic profiles 1.21.1 and 1.23.1 appear to be identical to allelic profile 1.3.1 (panel A. and C).