Abstract
Scientific collaborations involving multiple institutions are increasingly commonplace. It is not unusual for publications to have dozens or hundreds of authors, in some cases even a few thousands. Gathering the information for such papers may be very time consuming, since the author list must include authors who made different kinds of contributions and whose affiliations are hard to track. Similarly, when datasets are contributed by multiple institutions, the collection and processing details may also be hard to assemble due to the many individuals involved. We present our work to date on automatically generating author lists and other portions of scientific papers for multi-institutional collaborations based on the metadata created to represent the people, data, and activities involved. Our initial focus is ENIGMA, a large international collaboration for neuroimaging genetics.
Keywords: semantic metadata, semantic science, neuroinformatics
1 Introduction
Significant scientific effort is devoted to describing data with appropriate semantic metadata. Many communities have data repositories that use semantic markup to describe datasets, which enables users to query the repositories to retrieve data based on metadata properties of interest. In neuroimaging, which is the focus of this work, neuroinformatics repositories exist (e.g., http://nitrc.org) where researchers may download images corresponding to subjects of a certain age range. However, this metadata has been used in very limited ways beyond the repositories. Once the datasets are extracted from a repository, they often become separated from their metadata when they are analyzed in a separate system. Published articles include citations to the datasets that contain a unique identifier provided by the original repository, however their original semantic metadata is not passed on and is only informally described in the articles.
We are interested in the use of semantic metadata to automatically generate portions of scientific papers that describe datasets included in the publication. For example, biomedical papers that use datasets collected from a group of study participants often include demographic tables with information that often exists in the metadata for the datasets (e.g., age ranges, clinical characteristics). Similarly, metadata for all the datasets used may include pointers to the people who collected the data, which would be included as authors of the paper. In large multi-institutional collaborations where papers include dozens or hundreds of authors, generating the author list by hand can be very tedious.
This paper presents our approach to generating portions of scientific papers based on a semantic repository of project information. We created a semantic repository of projects, contributors and datasets, and used it to automatically generate author lists and descriptions of datasets. We use the Organic Data Science framework that we developed in prior work [2], which extends the Semantic MediaWiki platform, and captures entities and properties in RDF while providing users with a very simple user interface. We are working with the ENIGMA (Enhancing Neuro Imaging Genetics through Meta-Analysis) Consortium [7], a neuroscience collaboration where projects span many contributors from different institutions around the world (http://enigma.usc.edu).
In other work, we developed an approach to automatically generate the methods sections of papers from scientific workflows and their associated metadata [1]. That work focused on generating descriptions of the computational steps involved in analyzing data. The work presented here is complementary, in that we show how additional portions of a scientific paper can be automatically generated.
The paper starts with a description of what kinds of portions of papers could be generated automatically from a semantic repository of project information. We also describe the ontology and semantic repository that we developed to represent the information for ENIGMA. We then present our approach to generate portions of scientific papers, and show a detailed example of a representative ENIGMA paper.
2 Complex Project Information: The ENIGMA Collaboration and Publications
To illustrate the potential uses of semantic metadata to automatically generate portions of papers, we use several examples of publications by the ENIGMA Consortium, which involves many international research groups.
Author lists are often organized based on the roles and contributions made to the work. Consider [3] with dozens of authors, or [4] with hundreds of authors. At some point an individual is tasked with the daunting task of assembling a complete author list, generate an ordering, compiling all their affiliations, and entering them into a journal’s database for manuscript submission.
The manuscripts themselves include tables with information about the datasets used. In ENIGMA, papers often report clinical associations with medical (brain) image features, pooled from dozens of individual imaging studies around the world. It is therefore typical to include tables of the data collected in each study (a cohort), or the details of the data image collection process (an acquisition protocol). These tables include the information for the brain scanner used, demographics of the cohorts involved and the inclusion and exclusion criteria for data in each study cohort. Other tables may include other data summaries, such as genotyping platforms, or diagnostic scales that may be more specific to a particular project. These tables provide important provenance information, and may not be identical across clinical focus areas. Table 1 shows an excerpt of a table of acquisition protocols for [5].
Table 1.
Excerpt of a table of acquisition protocols of cohorts for [5].
| Site | Sequence | Field Strength | Acquisition Direction | # of Slices | Slice Gap | Voxel Size (mm3) | TI | TE | TR | Flip Angle | Citation | Segmentation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Amsterdam | 3D T1-weighted turbo field echo (TFE) | 3T scanner Philips Gyroscan Intera | coronal | 182 | 0mm | 1×1×1.2 | 0ms | 4.6ms | 9.621ms | 8 | 1, 2 | FreeSurfer (5.0) |
| Barcelona (Site 1: FIDMAG) | 3D T1-weighted enhanced fast gradient echo (EFGRE3D) | 1.5T GE Signa | Axial | 180 | 0mm | 0.47×0.47×1 | 710ms | 3.93ms | 2000ms | 15 | 3, 4 | FreeSurfer (5.3.0) |
Gathering information about authors and study metadata for papers from multi-institutional collaborations is a very tedious process. Authors come from numerous institutions around the world, and each may have multiple institutional and department affiliations. Almost 300 authors and 200 institutions are listed in [4]. To add to the complexity, journals often require information on author contributions and each author may contribute to one or more aspects of the project. Keeping track of who did what can get quite difficult, particularly since some authors (e.g., students) may have left research or changed institution by the time the manuscript is compiled. The information about datasets must be gathered from each participating cohort. Gathering information may become very time consuming as each cohort may have recorded this information in a different manner, yet the table provided for the manuscript must have somewhat consistent entries across all cohort datasets involved.
The examples given here are representative data-rich aspects of manuscripts that may be automatically generated. Although we focus on ENIGMA, large scale collaborations are becoming a key aspect of data discovery in the biomedical and broader scientific research fields. Our requirements are shared by large multi-institutional scientific collaborations, such as the climate collaboration described in [6].
3 A Semantic Repository of Complex Project Information for ENIGMA
This section describes the ontology that we created to describe large multi-institutional collaborations and its use in a semantic repository for ENIGMA.
3.1 A Scientific Collaboration Ontology
The collaboration ontology for the ENIGMA consortium was created to represent information that is crucial to organize the different activities, datasets and contributors. The classes and properties were created to represent information that is important to show in the manuscripts that result from collaborative activities. We created an initial collaboration ontology to fit the needs of ENIGMA.
Figure 1 shows the main concepts of the ontology, which include Working Group, Project, Dataset (collected for a group of people or cohort), Acquisition Procedure (called Protocol), and Person. It also shows the relationships between them. While some ontologies focus on people and projects and others focus on describing datasets, this ontology combines both aspects of the collaboration. We extended this core ontology with classes and properties that are used to describe projects, datasets, and people in ENIGMA. The ontology could be extended similarly for other collaborations by creating suitable properties of datasets and acquisition procedures in their particular domain.
Figure 1.

Core classes of the collaboration ontology.
To generate the author list, the contributors of the project, along with their role for that particular project, must be known. Thus, the “hasPSeniorLead,” “hasPJuniorLead,” and “hasPSpecialContributor” properties of the Project class allow the system to find out who needs to be acknowledged in the author list. In addition to the leadership roles of the project, the people who took charge of the cohorts are also acknowledged. Therefore, the Cohort class has two properties that represents who the principal investigator and the other investigators: “hasPI” and “hasInvestigator.”
A table with image acquisition protocols is necessary to include in all manuscripts that include imaging data, to describe the data collection procedure for each cohort. The image acquisition protocol table currently varies by paper but the most common columns were made into properties in the ontology. The “hasAcquisitionProcedure” property relates an image acquisition protocol to its respective cohort. The “AcquisitionProcedure” class has a “ImageAcquisitionProtocol” subclass with the following properties which are used to generate the image acquisition protocol table: “hasAcquisitionDirection,” “hasSequence,” “hasScanner,” “hasDataAcquisitionMatrix,” “hasFlipAngle,” “hasFoV,” “hasNumberOfEchoes,” “hasNumberOfSlices,” “hasScanTime,” “hasSliceThickness,” and “hasVoxelSize.”
A demographics table is common for all manuscripts that study human populations, and therefore key components of ENIGMA papers. Datasets can be collected from a cohort, i.e., a group of people who participate in a study. To find the total number of participants in a cohort, the number of males, and the number of females of a cohort, one can use the “hasNumberOfParticipants,” “hasNumberOfMales,” and “hasNumberOfFemales,” respectively. Since it is important to describe separately the group of patients and the group of controls in a study cohort, a new class called “CohortGroup” was created. Thus, a Cohort has the properties “hasControlGroup” and “hasDiagnosticGroup” with “CohortGroup” as their range. The “CohortGroup” has a “hasNumberOfParticipants,” “hasNumberOfMales,” and “hasNumberOfFemales” as properties, which are necessary to generate the cohort demographics table.
An inclusion and exclusion table is also part of a typical ENIGMA paper, as it is a key aspect of many clinical studies that study a particular patient population. The inclusion and exclusion criteria are described through a series of properties for the Cohort class. The criteria are not properties of the Cohort class as the control group and diagnostic group of a cohort can have different inclusion and exclusion criteria. Thus, separating the inclusion and exclusion criteria for the patient group and the control group was necessary. The “CohortGroup” properties that begin with “has” are inclusion properties while the “CohortGroup” properties that begin with “didNot” or “doesNot” are exclusion properties. The following properties are properties that describe the inclusion criteria for a specific cohort group: “hasDisorder,” “hasDisorderDetails,” “hasFirstDegreeRelativeWithDisorder,” “hasFirstEpisodeOf,” “isWithin,” “hasMedicalRatingDetails,” “usesTreatment,” “hasTreatmentDetails,” “hasNeurologicalComorbidity,” “hasPsychiatricComorbidity,” “isClinicallyStable,” and “isProficientinLocalLanguage.” The following properties describe exclusion criteria: “didNotHaveFirstEpisodeOf,” “doesNotHaveDisorder,” “doesNotHaveFirstDegreeRelativeWithDisorder,” “doesNotUseTreatment,” “doesNotHaveContraindicationToMRI,” “doesNotHaveNeurologicalComorbidity,” “doesNotHavePsychiatricComorbidiy,” “excludesLeftHands,” “excludesRightHands,” “isNotPregnant,” and “doesNotHaveIntellectualDisability.”
Many of these properties have an inverse. For example, the property “hasSeniorLead” has domain project and range person, and “isSeniorLeadOfP” is its inverse. There are also additional properties that describe the ENIGMA concepts further. For example, a project has a “hasApprovedProposalForm” property that links to the proposal that the project leads originally submitted to describe the project and have it approved.
3.2 A Semantic Repository for the ENIGMA Collaboration
We use the Organic Data Science framework [2] for collecting and managing information about ENIGMA. The framework is built on Semantic MediaWiki, and represents objects and properties in RDF. Users are shown all the properties relevant to the class of a resource on a wiki page as a table, and fill out their values. Figure 2 shows an example of a wiki page for an acquisition protocol. Users may add new properties, as shown at the bottom of the figure.
Figure 2.
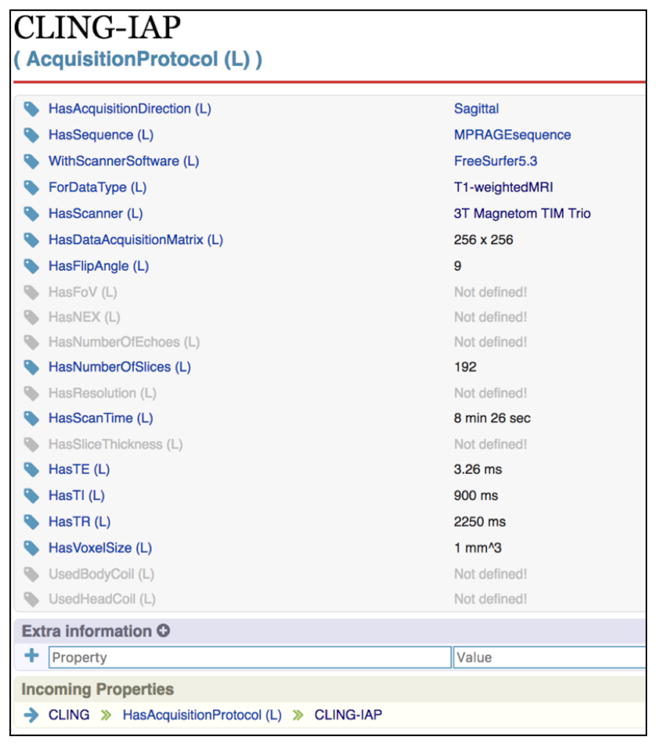
An example protocol page of the ENIGMA repository.
The ENIGMA repository is being prototyped with 3 selected projects out of more than 50 ENIGMA projects, with a total of 405 pages. It includes 3 working groups, 3 projects, 4 image acquisition protocol pages, 8 scanners, 89 cohort groups, 54 cohorts, and 112 persons. We continue to grow the repository so that all of the working groups, projects, cohorts, and researchers will be eventually represented. The repository is currently private to ENIGMA members.
4 Generation of Portions of Scientific Papers from a Semantic Repository
There are different approaches to organizing an author list. In ENIGMA, the following are two typical approaches. One approach is based on fine grained contribution, where the authors are ordered based on their roles. Under each role, the authors are alphabetically ordered. Another approach is based on coarse grained contribution, where the authors from roles other than junior and senior leads are placed in the middle of the list in alphabetical order. The junior and senior leads do not follow alphabetical ordering in either approach.
To generate the list of authors automatically, the system extracts the names and details of researchers involved in a project by leveraging the ENIGMA ontology. The list of authors is generated using one of the two approaches described above.
In addition to the author list, our system generates a detailed credit section that is often included in the acknowledgements of papers listing each individual’s contribution to the work. Authors who have multiple roles are credited for all of their roles.
The system automatically generates three types of tables: an imaging acquisition protocol table, a demographics table, and an inclusion/exclusion criteria table.
Table 2 shows an example of an automatically generated image acquisition protocol table. Image acquisition protocol tables display the information regarding the MRI scanner and imaging sequence used to scan each cohort. Here, the CLING acquisition protocol metadata from Figure 2 was used.
Table 2.
Generated image acquisition protocol table for CLING and HMS.
| Cohort | Data Type | Scanner | Acquisition Direction | Sequence | Data Acquisition Matrix | Flip Angle | Number of Slices | Scan Time | TE | TI | TR | Voxel Size |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CLING | T1-weighted MRI | 3T Magnetom TIM Trio | Sagittal | MPRAGE sequence | 256 × 256 | 9 | 192 | 8 min 26 sec | 3.26 ms | 900 ms | 2250 ms | 1 mm^3 |
| HMS | T1-weighted MRI | 1.5T Magnetom Sonata | Sagittal | MPRAGE sequence | 256 × 256 | 15 | 176 | 5 min | 4.0 ms | 700 ms | 1900 ms | 1 mm^3 |
Table 3 shows an example of an automatically generated demographics table. Demographics tables show the general information about the study participants in each cohort. Here, our table displays the cohort, diagnostic information (if applicable), the total number of individuals in each cohort broken down to the number of males and females, and the average age and age range of each group.
Table 3.
Generated demographics table for CLING and HMS.
| Cohort | Total | Control Total | Patient Total | Male Patients | Female Patients |
|---|---|---|---|---|---|
| CLING | 372 | 323 | 49 | 36 | 13 |
| HMS | 101 | 55 | 46 | 32 | 14 |
Inclusion/exclusion criteria tables include, for each cohort, information on any inclusion or exclusion criteria used for enrolment in a study. In MRI studies, individuals are excluded for having metal implants which may interfere and cause harm when placed inside a high magnetic field (i.e., MRI machine). Due to space constraints, we have omitted an example of this type of table.
5 Discussion
The author generation system assumes that separate pages are maintained for each author with details on their full name, their full set of affiliations, their highest degree, and their contact email (used only for a corresponding author). One limitation of the current application is that it assumes the authors are individuals and does not address cases where a consortium or group are included as authors. It also assumes that the author list always follows one of the two author list approaches described above, yet there can be many other possible approaches to author ordering. In addition, author information could be obtained from existing repositories such as ORCID or VIVO.
Currently our system can only generate three types of tables containing certain preselected columns. We envision custom table formats being created by project leads, so that they can easily be used by other projects within the ENIGMA consortium.
Significant amounts of information about ENIGMA are available in unstructured form. For example, the image acquisition protocols and the inclusion/exclusion criteria are not structured. The ENIGMA ontology allows for the metadata to be harmonized and reported in a standardized way. Now the ENIGMA repository has the potential to go beyond table generation for papers and allow filtering and selecting datasets by using standardized metadata. We believe these efforts will allow for improved collaborations and scientific discovery.
6 Conclusions
As scientific collaborations become more complex, documenting the details of the datasets used becomes increasingly challenging since it involves gathering information from dozens or hundreds of individuals across many institutions. We have shown that a semantic metadata repository for a collaboration enables the creation of tools to generate automatically author lists and tables that summarize key information about data collection and other data characteristics that are important to an article. We have an initial implementation of a semantic repository and associated generation tools for the ENIGMA neuroimaging genetics collaboration, which we continue to extend both in content and capabilities.
Acknowledgments
We are very grateful to the KAVLI foundation for their support of ENIGMA Informatics (PIs: Jahanshad and Gil). We also acknowledge support from the National Science Foundation under awards IIS-1344272 (PI: Gil), ICER-1541029 (Co-PI: Gil), and IIS-1344272 (PI: Gil), and from the National Institutes of Health’s Big Data to Knowledge Grant U54EB020403 for support for ENIGMA (PI: Thompson). We thank the members of the Organic Data Science and Linked Earth projects for their contributions to the design of the framework. We also thank the many participants of the ENIGMA collaboration for their feedback and other contributions to this work.
References
- 1.Gil Y, Garijo D. Towards Automating Data Narratives. Proceedings of the Twenty-Second ACM International Conference on Intelligent User Interfaces (IUI-17); 2017. [Google Scholar]
- 2.Gil Y, Garijo D, Ratnakar V, Khider D, Emile-Geay J, McKay N. A Controlled Crowdsourcing Approach to Scientific Ontology Development and Data Annotation. Proceedings of the Sixteenth International Semantic Web Conference (ISWC); 2017. [Google Scholar]
- 3.Guadalupe T, Mathias SR, vanErp TG, et al. Human subcortical brain asymmetries in 15,847 people worldwide reveal effects of age and sex. Brain Imaging and Behavior. 2016 doi: 10.1007/s11682-016-9629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hibar DP, Stein JL, Renteria ME, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–229. doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hibar DP, Westlye LT, Doan NT, Jahanshad N, et al. Cortical abnormalities in bipolar disorder: an MRI analysis of 6503 individuals from the ENIGMA Bipolar Disorder Working Group. Molecular Psychiatry. 2017 doi: 10.1038/mp.2017.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.PAGES2k Consortium. Emile-Geay Julien, McKay Nicholas P, et al. A global multiproxy database for temperature reconstructions of the Common Era. Scientific Data. 2017;4 doi: 10.1038/sdata.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, Toro R, Jahanshad N, et al. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging and Behavior. 2014;8(2) doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


