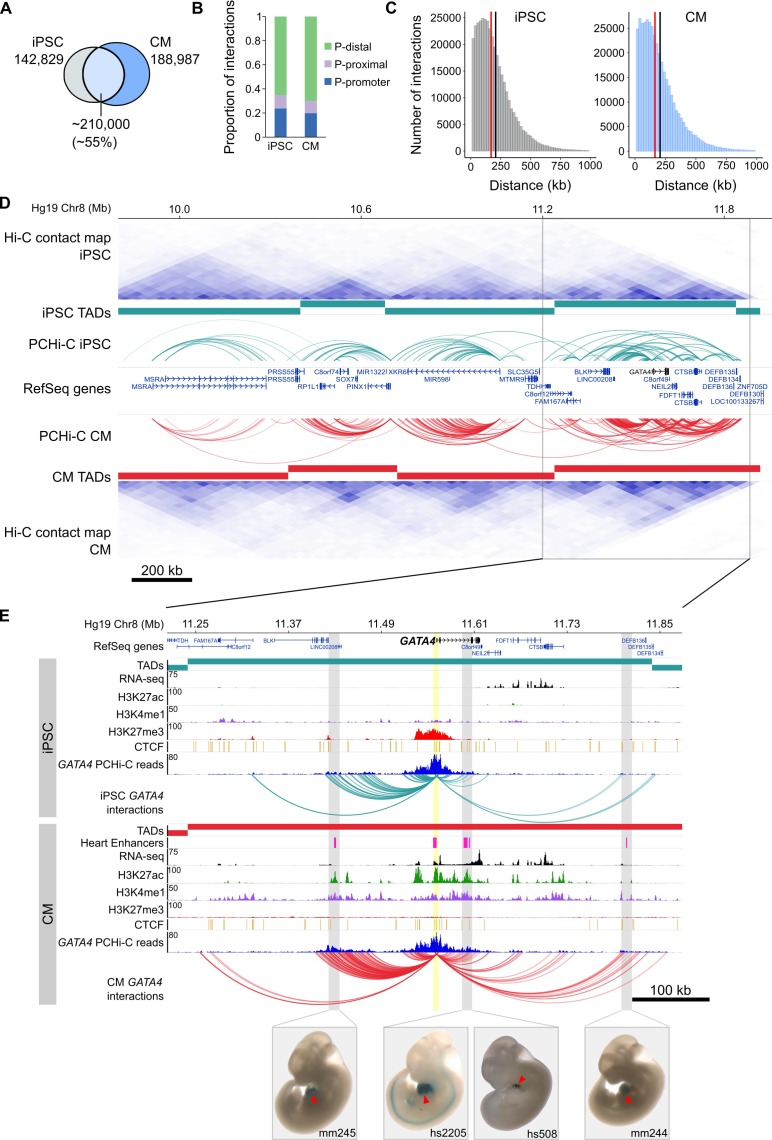
Figure 1. General features of promoter interactions.
(A) Venn diagram displaying the number of cell-type-specific and shared promoter interactions in each cell type. (B) Proportion of interactions in each distance category: promoter (P)-promoter (both interacting ends overlap a transcription start site (TSS)); P-proximal (non-promoter end overlaps captured region but not the TSS); P-distal (non-promoter end is outside of captured region). Note that all promoter interactions are separated by at least 10 kb. (C) Distribution of the distances spanning each interaction in iPSCs and CMs. The red line depicts the median (170 kb in iPSCs, 164 kb in CMs); the black line depicts the mean (208 kb in iPSCs, 206 kb in CMs). (D) A ~ 2 Mb region of chromosome 8 encompassing the GATA4 gene is shown along with pre-capture (whole genome) Hi-C interaction maps at 40 kb resolution for iPSCs (top) and CMs (bottom). TADs called with TopDom are shown as colored bars (median TAD size = 640 kb in both cell types, mean TAD size = 742 kb in iPSCs and 743 kb in CMs) and significant PCHi-C interactions as colored arcs. (E) Zoomed-in view of the GATA4 locus (promoter highlighted in yellow) in iPSCs (top) and CMs (bottom) along with corresponding RNA-seq data generated as part of this study, and ChIP-seq data for H3K27ac, H3K4me1, H3K27me3 and CTCF from the Epigenome Roadmap Project/ENCODE (H1 and left ventricle for iPSC and CM, respectively). Filtered GATA4 read counts used by CHiCAGO are displayed in blue with the corresponding significant interactions shown as arcs. For clarity, only GATA4 interactions are shown. Gray highlighted regions show interactions overlapping in vivo validated heart enhancers (pink boxes), with representative E11.5 embryos for each enhancer element (Visel et al., 2007). Red arrowhead points to the heart.