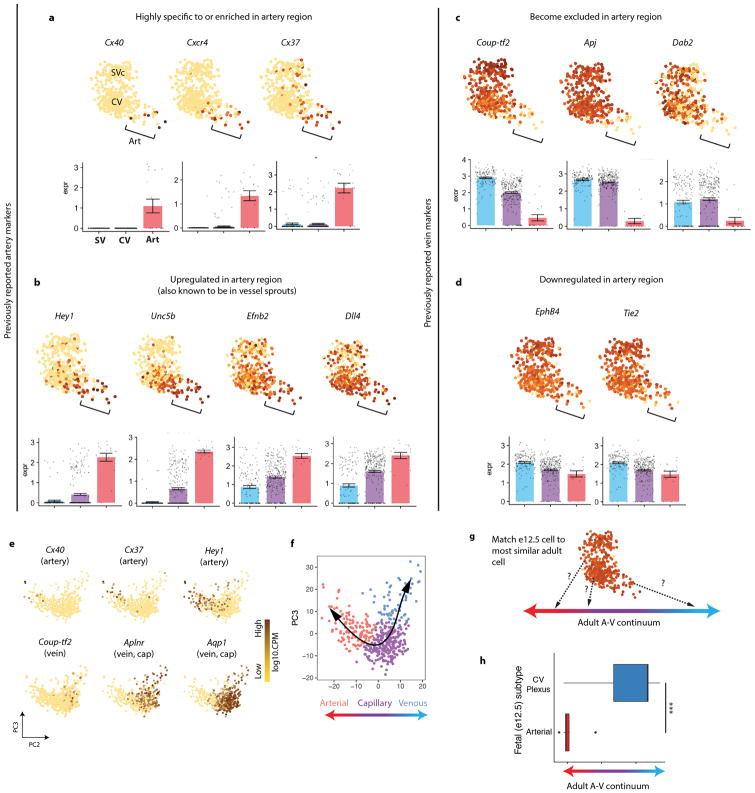
Extended Data Fig. 3. Characterization of pre-artery cells.
(a–d) rPCA plots of the e12.5 SVc-coronary vessel continuum. Each dot is an individual cell, and gene expression levels are indicated by the color spectrum as shown in Fig. 1d, which reflects log10 of counts per million. (a) Arterial genes highly enriched in the arterial areas of the plot. (b) Arterial genes significantly upregulated in, but not specific to, the arterial area of the plot. (c) Venous genes highly depleted in the arterial areas of the plot. (d) Venous genes downregulated, but not depleted, in the arterial area of the plot. For a–d, Bonferroni-adjusted p < 0.01; PCA plots, n=415 cells. Center and error bars are mean ± s.e.m. of log cpm expression values. (e) Genes expressed in adult coronary artery cells. Data is from the Tubula Muris consortium. N=445 cells. (f) Assignment of artery, capillary, and vein in adult coronary cells based on gene expression enrichment in e. N=445 cells. (g) Schematic for comparing e12.5 coronary cells to those along the adult artery-capillary-vein continuum. (h) Results of experiment schematized in g. The center line correspond to the median; the upper and lower hinges correspond to the first and third quartile, respectively; the whiskers extend to the largest value or to 1.5*IQR (inter-quartile range, or distance between quartiles), whichever is smaller. Pre-artery cells: n=20 cells. CV: n=277 cells. P = 6.2 x 10−13. Statistical test is two-tailed. A, artery; Art, arterial; CV, coronary vessel plexus; PC, principle component; SVc, sinus venosus-coronary; V, vein