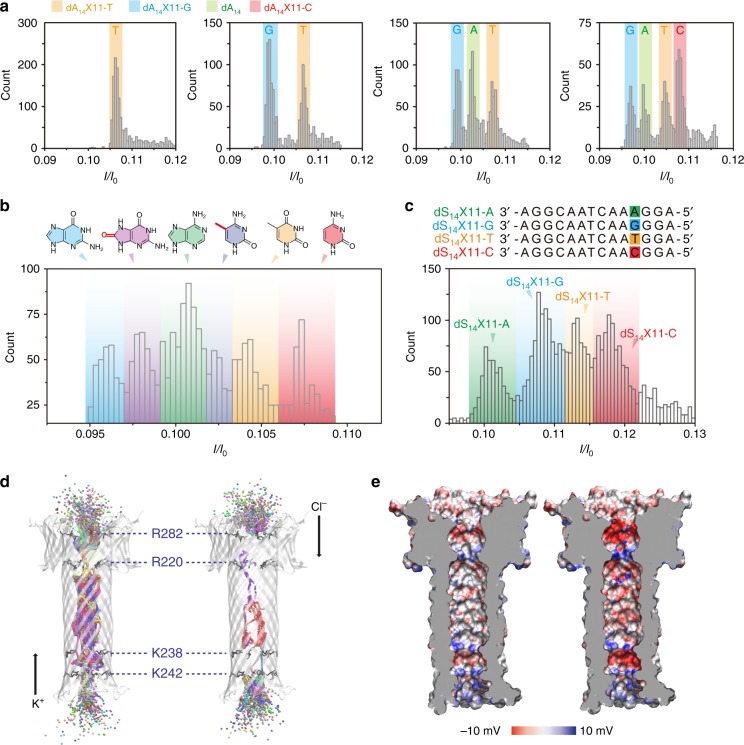
Fig. 4.
Discrimination of all four DNA bases by dA14X11 sites of the aerolysin nanopore. a The I/I0 histograms of dA14X11-T; dA14X11-T and dA14X11-G; dA14X11-T, dA14X11-G, and dA14; dA14X11-T, dA14X11-G, dA14, and dA14X11-C, from left to right. The number of blockades in each histogram is at least 2000. b The I/I0 histograms for aerolysin pore interrogated with six kinds of oligonucleotides that only differ at the position of 11 (four kinds of normal base: A, T, C, G and two kinds of damage nucleobases: mC and oG). The number of blockades is 4500. c Histograms of I/I0 caused by the four kinds of nucleobase in a heteropolymeric DNA strand. The sequence information of four ssDNA is illustrated at the top of the figure. The data were obtained for 3.0 M KCl, 10 mM Tris, and 1.0 mM EDTA, pH=8.0, 17 ± 2 °C at the bias potential of +100 mV. The number of blockades in each histogram is at least 5000. d Potassium (left) and chloride (right) ion occupancy at each 0.1 ns of the MD simulation. The DNA (dA14) is not shown for sake of clarity. Ions are represented as spheres colored according to residue ID. Amino acids R282, R220, K238, and K242 are shown as black sticks. e Electrostatic surface of aerolysin pore lumen as calculated with the APBS-PDB2PQR server59,60, without (left) or with (right) dA14 inserted in the pore. The DNA is not shown for the sake of clarity