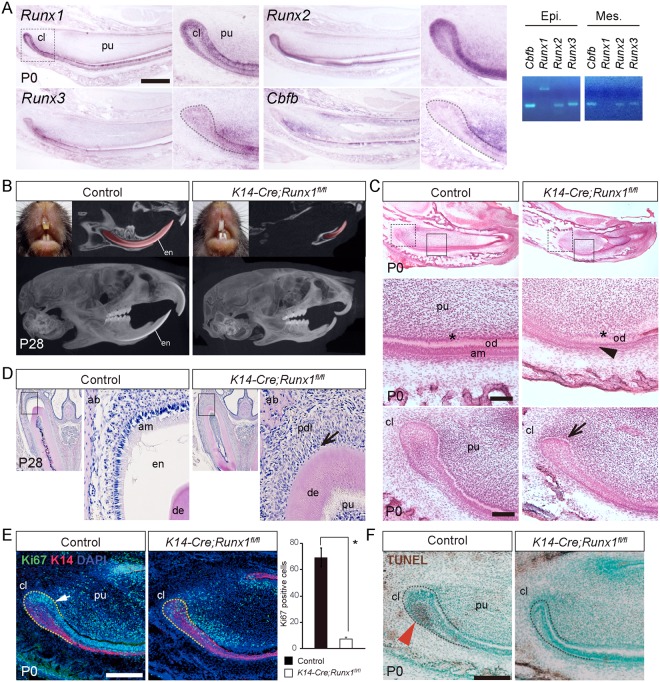
Figure 1.
The incisor phenotype of K14-Cre/Runx1fl/fl mice. (A) Expression of Runx1, Runx2, Runx3 and Cbfb in sagittal sections of P0 incisors and gene expression was confirmed by RT-PCR from dissected epithelium and mesenchyme. Runx1 transcripts were detected in the dental epithelium. In the cervical loop, Runx1 was expressed in the basal epithelial cells (inner enamel epithelium) and the outer enamel epithelium in the cervical loop. Scale bar: 500 μm. (B) The Runx1 mutant incisors demonstrated a column shape and lost their transparency, although the control incisors developed fine edged shape. In the micro-CT analysis, the incisors of the Runx1 mutant mice had an abnormal shape, extreme shortening and enamel defects. On the other hand, significant differences were not detected in the molar root length or morphology. (C) HE staining of incisor sagittal sections from P0 Runx1 mutant mice confirmed the shortening of the Runx1 mutant incisors. At higher magnification, hypoplasia of the cervical loop (arrow) was evident and ameloblast precursors lost their polarity and had a flattened shape in Runx1 mutants (arrowhead). Scale bar: 100 μm. (D) HE staining of incisor frontal sections from P28 Runx1 mutant mice confirmed the lack of enamel matrix formation. At higher magnification, enamel matrix was not evident between the dentin and the periodontal ligament (arrow) in the upper incisors of Runx1 mutants. (E) Double immunostaining for Ki67 (green) and K14 (red) revealed that the epithelium in the cervical loop of the Runx1 mutant was significantly less proliferative (arrow). Nuclei were counterstained with DAPI (blue). Scale bar: 200 μm. (F) TUNEL staining revealed that the number of TUNEL-positive cells was significantly reduced at the cervical loop of the Runx1 mutant incisors (arrowhead). cl, cervical loop; pu, dental pulp; en, enamel; am, ameloblasts; od, odontoblasts. Scale bar: 200 μm.