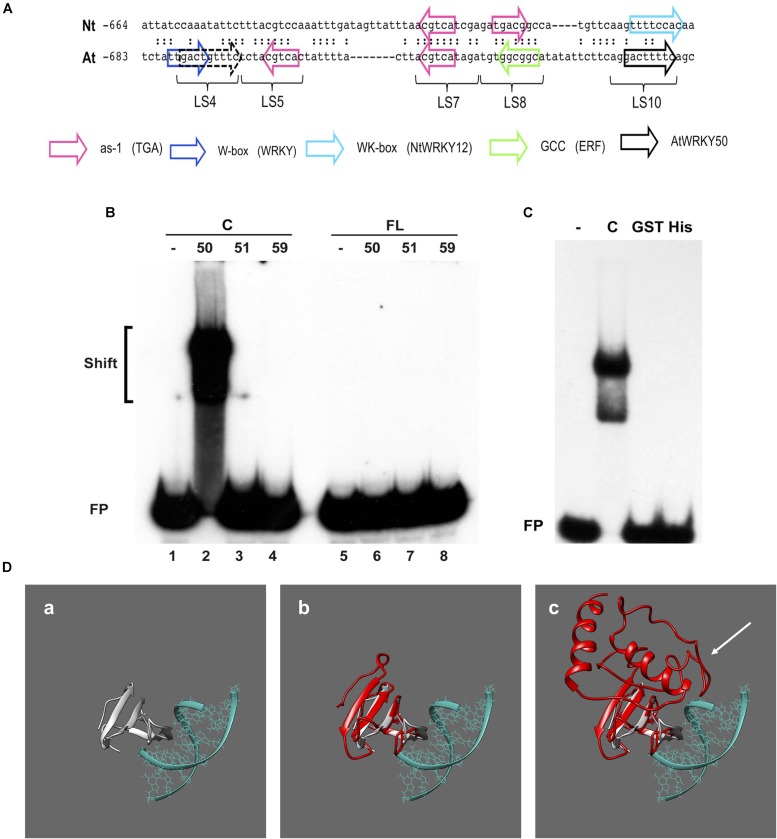
FIGURE 3.
AtWRKY50 binds to the PR1 promoter. (A) Comparison of sequences in the promoters of tobacco PR1a (Nt) and Arabidopsis PR1 (At). Only the sequence of the top strands is given. The sequences of the promoter regions are shown with gaps to allow maximal alignment. The position of the leftmost nucleotide relative to the transcription start site is indicated. Corresponding nucleotides are indicated by colons. Colored block arrows mark consensus binding sites for various transcription factors, as indicated. The direction of the arrow indicates whether the consensus sequence is in the top (right-pointing arrow) or bottom strand. The dashed and solid black arrows mark the binding sites for AtWRKY50. The positions of sequence elements used in the linker scanning analysis of the PR1 promoter by Lebel et al. (1998) are indicated (LS). (B) EMSAs were performed with an 80-bp fragment of the PR1 promoter and GST-tagged C-terminal halves (Lanes C) or full-length (Lanes FL) versions of AtWRKY50, -51 and -59, as indicated above the lanes. (C) EMSAs were performed with the same probe together with the GST-tagged C-terminal half (Lane C) and GST-tagged (Lane GST) and His-tagged (Lane His) full-length versions of AtWRKY50. In (B,C), lanes labeled with the minus sign were loaded with the probe only. The positions of shifts and the unbound probe (FP) are indicated. (D) Modeling prediction analysis of C-terminal and full-length AtWRKY50 protein binding to DNA in silico. (a) shows the structure of C-terminal WRKY4-C domain bound to DNA from Yamasaki et al. (2012) onto which AtWRKY50 and its truncated form are superimposed. The overlap of the β-sheets and overall structure correlating with interactions is shown in (b). The full length AtWRKY50 sequence also demonstrates a superimposable C-terminal region (c) but the additional protein structure was predicted to possess a large disordered region (shown with white arrow).