FIGURE 4.
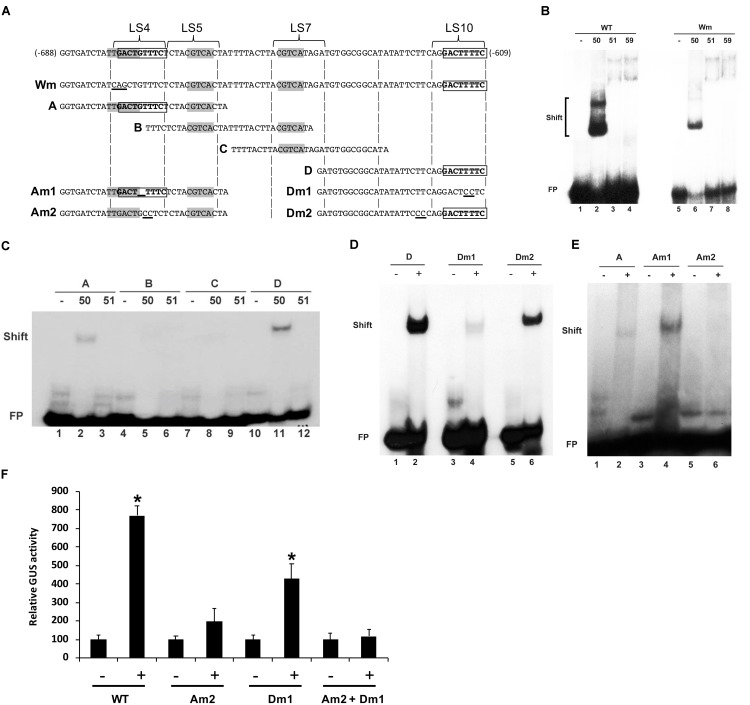
Characterization of AtWRKY50’s binding sites on the PR1 promoter (A) Sequences of PR1 promoter fragments used for EMSAs. Only the sequence of the upper strand is given. The top line displays the sequence of the 80-bp fragment corresponding to bp –688 to –609 upstream of the transcription start site. Regions LS4, LS5, LS7, and LS10, as used in the linker scanning analysis of Lebel et al. (1998), are indicated. Wm indicates an 80-bp fragment with a mutation (TTGACT to TCAGCT) in the W-box in LS4. Overlapping subfragments A, B, C, and D, and their mutant versions Am1, Am2, Dm1 and Dm2 are aligned with the sequence of the 80-bp fragment. The W-box (TTGACT) and the CGTCA boxes of the as-1 element are indicated in bold. Mutations in Wm, Am1, Am2, Dm1, and Dm2 are underlined. (B) EMSAs were performed with wild type 80-bp PR1 promoter fragment (WT) or with an 80-bp fragment with a mutation in the W-box (Wm) as probes together with the GST-tagged C-terminal halves of AtWRKY50, -51 and -59, as indicated above the lanes. The positions of band shifts and the unbound probe (FP) are indicated. Lanes labeled with the minus sign were loaded with probe only. (C) AtWRKY50 binds to the PR1 promoter at two positions. EMSAs were performed with overlapping PR1 promoter fragments A, B, C, and D as probes and GST-tagged AtWRKY50-C or the C-terminal half of AtWRKY51, as indicated above the lanes. The positions of band shifts and the unbound probe (FP) are indicated. Lanes labeled with the minus sign were loaded with probe only. (D,E) AtWRKY50 binds to the LS10 and LS4 element in the PR1 promoter. EMSAs were performed with wild type (D) and WT (A) and mutant versions (Dm1, Dm2) of fragment D and Am1, Am2 of fragment A of PR1 promoter as probes and GST-tagged AtWRKY50-C, as indicated above the lanes. The positions of band shifts and the unbound probe (FP) are indicated. Lanes labeled with the minus sign were loaded with probe only; lanes labeled with the plus sign were loaded with the probe and AtWRKY50-C. (F) PR1 activation by AtWRKY50 requires intact binding sites. Arabidopsis protoplasts were co-transfected with WT and mutant PR1::GUS construct alone (minus sign) or together with expression plasmids 35S::AtWRKY50. After incubation GUS activity was measured spectrophotometrically. Expression levels (%) are given relative to expression level without WRKY effector. Statistical differences among the samples is labeled with asterisk (p < 0.05).