Abstract
Background
Antibiotic-associated gastrointestinal signs (AAGS) occur commonly in cats. Co-administration of synbiotics is associated with decreased AAGS in people, potentially due to stabilization of the fecal microbiome and metabolome. The purpose of this double-blinded randomized-controlled trial was to compare AAGS and the fecal microbiome and metabolome between healthy cats that received clindamycin with a placebo or synbiotic.
Methods
16 healthy domestic shorthair cats from a research colony were randomized to receive 150 mg clindamycin with either a placebo (eight cats) or commercially-available synbiotic (eight cats) once daily for 21 days with reevaluation 603 days thereafter. All cats ate the same diet. Food consumption, vomiting, and fecal score were recorded. Fecal samples were collected daily on the last three days of baseline (days 5–7), treatment (26–28), and recovery (631–633). Sequencing of 16S rRNA genes and gas chromatography time-of-flight mass spectrometry was performed. Clinical signs, alpha and beta diversity metrics, dysbiosis indices, proportions of bacteria groups, and metabolite profiles were compared between treatment groups using repeated measures ANOVAs. Fecal metabolite pathway analysis was performed. P < 0.05 was considered significant. The Benjamini & Hochberg’s False Discovery Rate was used to adjust for multiple comparisons.
Results
Median age was six and five years, respectively, for cats in the placebo and synbiotic groups. Hyporexia, vomiting, diarrhea, or some combination therein were induced in all cats. Though vomiting was less in cats receiving a synbiotic, the difference was not statistically significant. Bacterial diversity decreased significantly on days 26–28 in both treatment groups. Decreases in Actinobacteria (Bifidobacterium, Collinsella, Slackia), Bacteriodetes (Bacteroides), Lachnospiraceae (Blautia, Coprococcus, Roseburia), Ruminococcaceae (Faecilobacterium, Ruminococcus), and Erysipelotrichaceae (Bulleidia, [Eubacterium]) and increases in Clostridiaceae (Clostridium) and Proteobacteria (Aeromonadales, Enterobacteriaceae) occurred in both treatment groups, with incomplete normalization by days 631–633. Derangements in short-chain fatty acid, bile acid, indole, sphingolipid, benzoic acid, cinnaminic acid, and polyamine profiles also occurred, some of which persisted through the terminal sampling timepoint and differed between treatment groups.
Discussion
Cats administered clindamycin commonly develop AAGS, as well as short- and long-term dysbiosis and alterations in fecal metabolites. Despite a lack of differences in clinical signs between treatment groups, significant differences in their fecal metabolomic profiles were identified. Further investigation is warranted to determine whether antibiotic-induced dysbiosis is associated with an increased risk of future AAGS or metabolic diseases in cats and whether synbiotic administration ameliorates this risk.
Keywords: Antibiotic-associated gastrointestinal signs, Short-chain fatty acids, Bile acids, Metabolome, Indole, Polyamines, Dysbiosis, Sphingolipids, Antibiotic-associated diarrhea, Probiotic
Introduction
Antibiotic administration is associated with profound, and sometimes prolonged, derangements of the fecal microbiome and metabolome of people and animals (De La Cochetière et al., 2010; Dethlefsen et al., 2008; Jakobsson et al., 2010; Suchodolski, 2016b; Suchodolski et al., 2009). This dysbiosis is believed to be a primary contributor to the development of antibiotic-associated gastrointestinal signs (AAGS), such as hyporexia, vomiting, and diarrhea (Hempel et al., 2012; McFarland, 2008; Videlock & Cremonini, 2012). Susceptibility to AAGS and derangements in the microbiome both increase with repeated antibiotic exposure (Ouwehand et al., 2016).
Antibiotic-associated gastrointestinal signs occur commonly in cats (Albarellos & Landoni, 2009; Albarellos & Landoni, 1995; Hunter et al., 1995) and people (Hempel et al., 2012; Lenoir-Wijnkoop et al., 2014; McFarland, 2008), and they are an important cause of antibiotic non-compliance (Chan et al., 2012; Jefferds et al., 2002; Kardas et al., 2005; Llor et al., 2013; Muñoz et al., 2014; Pechere et al., 2007). Co-administration of probiotics or synbiotics (commercial mixtures of probiotics and prebiotics) with antibiotics is associated with up to a 3-fold decrease in AAGS in people (Hempel et al., 2012; Johnston et al., 2011; Lenoir-Wijnkoop et al., 2014; Narayan et al., 2010; Selinger et al., 2013). Positive effects of probiotics on AAGS in people are postulated to result from stabilization of the fecal microbiome and metabolome (Hempel et al., 2012; Ouwehand et al., 2016).
The impact of synbiotics on AAGS and antibiotic-induced dysbiosis in cats is currently unclear. However, the administration of one commercially-available multi-strain synbiotic (Proviable-DC; Nutramax Laboratories Veterinary Sciences, Inc., Lancaster, SC, USA) resulted in significantly improved fecal scores for 72% of cats with idiopathic chronic diarrhea (Hart et al., 2012), suggesting potential efficacy for reducing gastrointestinal signs due to other causes. Prevention or mitigation of AAGS could decrease noncompliance by clients and, thus, patient morbidity. Even if direct clinical effects are not observed, synbiotic administration might be warranted if it is found to decrease antibiotic-induced changes in the microbiome and metabolome and, thus, potentially reduce cumulative long-term sensitivity to AAGS and/or metabolic disorders associated with antibiotic exposure, such as inflammatory bowel disease and obesity (Aniwan et al., 2018; Kronman et al., 2012; Ouwehand et al., 2016).
The purpose of this study was to compare changes in food intake, vomiting, fecal scores, the fecal microbiome, and the fecal metabolome of healthy cats receiving either a placebo or a commercially-available synbiotic with 150 mg clindamycin orally once daily for 21 days. A secondary objective of the study was to assess the long-term impacts of antibiotic and synbiotic administration on the fecal microbiome and untargeted metabolomic profiles of healthy cats.
Materials and Methods
Study population
This study was performed at the University of Tennessee’s Veterinary Medical Center and was approved by the Institutional Animal Care and Use Committee at the University of Tennessee, Knoxville (protocol number 2169). Twenty purpose-bred, domestic short hair cats from the University of Tennessee, College of Veterinary Medicine teaching and research colony determined to be healthy based on unremarkable physical examinations at the time of study initiation and lack of pertinent abnormalities on CBC or biochemistry profiles performed within the previous six months were enrolled. Prior antibiotic use was not noted in any of the medical records. All cats ate the same commercial adult dry cat food for the duration of the study (Hill’s Science Diet Light; Hill’s Pet Nutrition, Inc., Topeka, KS, USA). Cats were transitioned to individual housing the week prior to the start of the study and randomized into 1 of 2 treatment groups (placebo vs. synbiotic) at the start of the study using a random number sequence.
Study periods
The study consisted of a baseline period (days 1–7), a treatment period (days 8–28), and a long-term follow-up period (days 631–633) (Fig. 1). During the baseline period, no treatments were administered. During the treatment period, all cats received 150 mg (34 mg/kg/d, range 25.9–44.1 mg/kg/d) of clindamycin (Antirobe Antirobe Aquadrops; Pfizer, Pharmacia & Upjohn Company, New York, NY, USA) PO once daily with food for 21 days. The dose was chosen based on the manufacturer drug insert (Antirobe Antirobe Aquadrops manufacturer insert, Pfizer Pharmacia, March 2010), which noted changes in fecal consistency and emesis in cats receiving higher doses of clindamycin. Cats received either one capsule of placebo or a commercially-available synbiotic PO once daily at the time of antibiotic administration. The synbiotic contains five billion cfu of a proprietary mixture of Bifidobacterium bifidum, Enterococcus faecium and thermophilus, and Lactobacillus acidophilus, bulgaricus, casei, and lantarum per capsule, as well a proprietary blend of fructooligosaccharide and arabinogalactan. The time period for treatment was chosen based on previous veterinary publications (Garcia-Mazcorro et al., 2011; Hart et al., 2012; Koeppel et al., 2006), which showed significant changes in the fecal microbiome or fecal score by 21 days of treatment. After completion of treatment, cats were returned to group housing in the colony. On day 631, cats remaining in the teaching and research pool were reevaluated. Based on review of their medical records, none of the cats had received antibiotics, probiotics, other medications known to affect the microbiome, or dietary change in the intervening time.
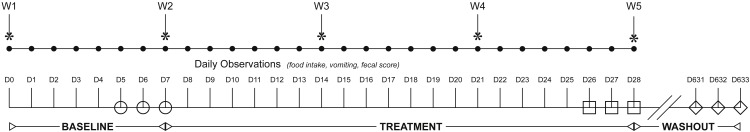
Figure 1. Flowchart illustrating study design, duration, observations, and sampling.
The study spanned 633 days (D1-633) and was broken into three study periods: baseline (D1–D7), treatment (D8–D28), and washout. Cats were randomized to receive either a placebo or synbiotic with 150 mg clindamycin PO once daily during treatment. Food intake, vomiting, and fecal score were recorded daily (•) and weight (W) weekly (*) by an individual blinded to treatment group. Two grams were collected from the center portion of each first morning naturally-voided fecal samples for each cat once daily for the last three days of each study period: baseline (open circles), treatment (open squares), and recovery (open diamonds). Each sample was subdivided into two aliquots, with each aliquot placed into a 2 mL cryovial and immediately frozen at −80 °C pending completion of data collection.
Data collection
All cats underwent weekly physical examinations, including determination of body weight, throughout the study. Daily observations, including food intake and vomiting (present vs. absent), were collected throughout the study by an observer (NLL) blinded to the treatment groups. Photographs of defecated feces were taken daily by the same observer. Fecal consistency was scored (Greco, 2011) using still photographs at the completion of data collection by two investigators (JCW and JES), who were blinded to the cat, treatment group, and day for each fecal sample.
Fecal samples
To limit the impact of daily variation in bacterial populations and fecal metabolites, as well as differential distribution of bacterial groups and metabolites within individual fecal samples, first morning naturally-voided fecal samples were collected daily for three days for each cat at each of three timepoints: the conclusion of baseline (days 5–7), the conclusion of treatment (days 26–28), and after long-term recovery (days 631–633). Two grams from the center portion of each fecal sample was subdivided into two aliquots, with each aliquot placed into a 2 mL cryovial. Samples were immediately frozen and remained in storage at −80 °C pending completion of data collection. Samples for each cat from each timepoint were combined directly prior to sample analysis to generate pooled samples for microbiome and metabolomic analysis.
Microbiome analysis
Genomic DNA was extracted from 100 mg of feces from each pooled sample using a commercially available kit according to manufacturer’s protocol (PowerSoil; Mo Bio, Carlsbad, CA, USA).
Amplification and sequencing of the V4 variable region (primers 515F/806R) of the 16S rRNA gene was performed on a MiSeq (Illumina; MR DNA (Molecular Research LP), Shallowater, TX, USA) as previously described (Bell et al., 2014). The software Quantitative Insights Into Microbial Ecology (QIIME, v. 1.8; http://www.qiime.org) was used for processing and analyzing the sequences. The raw sequence data were de-multiplexed, and low quality reads were filtered using default parameters for QIIME. Chimeric sequences were filtered from the reads using USEARCH against the 97% clustered representative sequences from the Greengenes database (v. 13.8) and removed. The remaining sequences were assigned to operational taxonomic units (OTUs) using an open-reference OTU picking protocol using the default QIIME parameters, uclust consensus taxonomy assigner, and Greengenes database. The OTU table was rarefied to 35,000 sequences per sample. Alpha rarefaction plots, alpha diversity metrics (Chao1, Shannon, Goods Coverage, and Observed Species), and beta diversity metrics (weighted and unweighted UniFrac distance matrices) were created using QIIME scripts.
Quantitative PCR was performed for selected bacterial groups (total bacterial, Faecalibacterium spp., Turicibacter spp., Streptococcus spp., Escherichia coli, Blautia spp., Fusobacterium spp., Clostridium hiranonis) using extracted DNA as has been previously described (AlShawaqfeh et al., 2017). Briefly, 2 µl of normalized DNA (final concentration: 5 ng/ µl) was combined with 5 µl of a DNA-binding dye (SsoFast EvaGreen supermix; Bio-Rad Laboratories, CA, USA), 0.4 µl each of a forward and reverse primer (final concentration: 400 nM), and 2.6 µl of PCR water to achieve a total reaction volume of 10 µl. Oligonucleotide primers and probes, as well as respective annealing temperatures, are summarized in Table 1. Data were expressed as log amount of DNA (fg) for each particular bacterial group per 10 ng of isolated total DNA.
Table 1. Target bacteria, oligonucleotide primers/probes, and annealing temperatures used for quantitative PCR analysis.
| Target | Primer type | Sequence (5′-3′) | Annealing (°C) |
|---|---|---|---|
| Universal bacteria | Forward | CCTACGGGAGGCAGCAGT | 59 |
| Reverse | ATTACCGCGGCTGCTGG | ||
| Blautia spp. | Forward | TCTGATGTGAAAGGCTGGGGCTTA | 56 |
| Reverse | GGCTTAGCCACCCGACACCTA | ||
| Clostridium hiranonis | Forward | AGTAAGCTCCTGATACTGTCT | 50 |
| Reverse | AGGGAAAGAGGAGATTAGTCC | ||
| Escherichia coli | Forward | GTTAATACCTTTGCTCATTGA | 55 |
| Reverse | ACCAGGGTATCTAATCCTGTT | ||
| Faecalibacterium spp. | Forward | GAAGGCGGCCTACTGGGCAC | 60 |
| Reverse | GTGCAGGCGAGTTGCAGCCT | ||
| Fusobacteria spp. | Forward | KGGGCTCAACMCMGTATTGCGT | 51 |
| Reverse | TCGCGTTAGCTTGGGCGCTG | ||
| Streptococcus spp. | Forward | TTATTTGAAAGGGGCAATTGCT | 54 |
| Reverse | GTGAACTTTCCACTCTCACAC | ||
| Turicibacter spp. | Forward | CAGACGGGGACAACGATTGGA | 63 |
| Reverse | TACGCATCGTCGCCTTGGTA |
Metabolomics analysis
10 mg of lyophilized feces from each pooled sample were analyzed at a metabolomics facility using gas chromatography time-of-flight mass spectrometry (GC-TOF-MS) and standardized protocols as described in detail elsewhere (Fiehn et al., 2008; Minamoto et al., 2015). Raw data files were processed using ChromaTOF v. 2.32. BinBase algorithm was applied to match spectra to database compounds (West Coast Metabolomics Core, University of California, Davis, CA, USA) or to characterize as an unknown compound as reported previously (Honneffer et al., 2017). Quantification was reported by peak height of an ion at the specific retention index characteristic of the compound across all samples.
Statistical analyses
Descriptive statistics were generated for each parameter. Samples were analyzed for normality using the Shapiro–Wilk test and, for the presence of outliers, using box-and-whisker plots. Age and weight for the two treatment groups were compared using an independent two-sample Student’s t-test. Mean percent food intake, percent days of vomiting, and mean fecal scores were determined for each week within each study period (baseline and treatment weeks 1, 2, and 3). Mean food intake for each week in each study period was calculated as a percentage of food intake of each cat during the week prior to the start of the study. Inter-rater correlation coefficients were calculated for fecal scores. The mean of fecal scores assigned by the two investigators were used for all further statistical analyses. Mean food intake, percent days vomiting per week, and mean fecal score were compared between treatment groups using a multivariate repeated measures ANOVA. Treatment group (placebo or synbiotic), week, and cat were included as categorical variables. Treatment, week, and the treatment-by-week interaction were included as fixed effects. Week was included as a repeated measure with subject as cat. Age, sex, and weight were initially included as covariates but were not retained in the final models. Model assumptions regarding normally distributed residuals were verified with the Shapiro–Wilk test for normality. Model assumptions regarding equality of variances were verified with Levene’s Test for Equality of Variances.
Global changes in microbiota communities (beta diversity) between individuals were determined using unweighted Unifrac distance metrics; principal coordinates analysis (PCoA) plots and rarefaction curves were plotted using QIIME software. The ANOSIM function in PRIMER 6 (PRIMER-E Ltd, Ivybridge, UK) was used to compare beta diversity metrics across time and between treatment groups (Suchodolski et al., 2012). A dysbiosis index was calculated through methods previously described (AlShawaqfeh et al., 2017). The dysbiosis index is a quantitative PCR-based assay that summarizes the quantitative abundances of 8 major bacterial groups in feces in 1 number. The higher the dysbiosis index, the more deviation of the microbiota from normobiosis. Global changes in untargeted metabolomic profiles were determined using principal component analysis (PCA) plots developed using an online metabolomics software analysis suite (MetaboAnalyst 3.0: http://www.metaboanalyst.ca) as per standard protocols (Xia et al., 2015; Xia & Wishart, 2011). Pathway analysis was performed in the same software suite using the Homo sapiens pathway library, interquantile range data filtering, log transformation, and Pareto scaling.
Shannon indices, goods coverage, the Chao 1 metric, the dysbiosis index, proportions of bacteria groups, and fecal metabolite profiles were compared between treatment groups using a mixed model, split-plot repeated measures ANOVA that included fixed effects of treatment (placebo or synbiotic), time period, and treatment-by-time period interaction. Cat nested within group was included as a random effect. Age and sex were included as covariates in the initial analysis, but they were not retained in the final model due to lack of significant effects. The repeated measure of time period was accounted for in a repeated statement. A compound symmetry variance/covariance structure was incorporated into each model to account for the potential inclusion of constant covariates over time. The Shapiro–Wilk test of normality of the residuals was evaluated for each marker to confirm the assumption of normally distributed residuals had been met. Model assumptions regarding equality of variances were verified with Levene’s Test for Equality of Variances. Differences in least squares means were determined for markers with significant main effect or interaction terms. Due to a marked lack of variability in microbiome results for cats on days 26–28 (see Results below), rank transformation had to be employed to allow convergence of the model for analysis of the proportions of bacteria groups and ensure the statistical assumptions regarding normally distributed residuals and equality of variances were met. Only bacteria taxa that were present in at least 50% of cats in ≥1 group at ≥1 time point were included in statistical analyses.
P < 0.05 was considered statistically significant. P-values were corrected for multiple comparisons on each phylogenetic level and for untargeted metabolomics using the Benjamini & Hochberg’s False Discovery Rate (fdr). Publicly-accessible software packages (http://www.qiime.org; http://www.metaboanalyst.ca; SAS 9.4 release TS1M3; MedCalc 15.8: MedCalc, Ostend, Belgium; SAS 9.4 release TS1M3: SAS Institute Inc., Cary, NC, USA) were used for all analyses.
Results
Study population
There were four female spayed and 6 male castrated cats initially enrolled in the placebo group and five female spayed cats and five male castrated cats in the synbiotic group. Median age was six years (range 3–8 years) for cats in the placebo group and five years (range 3–8 years) for cats in the synbiotic group. Median weight was 4.2 kg (range 3.4–5.4 kg) for cats in the placebo group and 4.3 kg (3.4–5.8 kg) for cats in the synbiotic group. No cat vomited any of the administered capsules during the study. Four cats were removed from the study for marked hyporexia (two placebo, one synbiotic) or high baseline fecal scoring (1 synbiotic).
After completion of treatment, cats were returned to the colony. On day 631, 15/16 of the cats that completed the trial were available for re-examination. One cat (from the synbiotic group) had been retired from the colony with subsequent private adoption. None of the cats had received antibiotics, probiotics, or other medications known to affect the microbiome in the intervening time, and none had undergone diet change. None of the cats were experiencing vomiting, abnormal fecal scores, or inadequate body condition scores.
Antibiotic-associated gastrointestinal signs
Food intake, percent days vomiting, and fecal scores over time by treatment group are summarized in Table 2. Food intake was decreased during treatment in 3/8 cats in the placebo group and 4/8 cats treated with the synbiotic. Food intake was significantly associated with week of treatment (F-value 5.2, P < 0.01) but did not differ between treatment groups (P = 0.5). Percent days vomiting per week was significantly associated with week of treatment (F-value 8.6, P < 0.01) but not treatment group (P = 0.4). It was highest in the first week of treatment, during which 14/16 cats had at least 1 episode of vomiting, seven in each group. Fecal scores increased significantly during treatment (F-value 9.4, P < 0.01) but did not differ between treatment groups (P = 0.2). All cats in each treatment group had fecal scores ≥5 at some point during treatment, although high daily variability was noted. There was no significant effect of age, weight, or sex on any of the clinical parameters evaluated.
Table 2. Antibiotic-associated gastrointestinal signs for cats that received clindamycin and a placebo or synbiotic.
Mean (± standard deviation) percent food intake compared to the week prior to initiation of the study, percent days vomiting per week, and mean fecal score for 16 healthy cats, eight per group, that received 150 mg clindamycin with either a placebo or synbiotic PO once daily for 21 days.
| Baseline | Treatment | |||
|---|---|---|---|---|
| Week 1 | Week 2 | Week 3 | ||
| Food intake (%) | ||||
| − Placebo | 97.8 ± 4.1 | 89.9 ± 5.5 | 89.7 ± 11.3 | 88.1 ± 13.2 |
| − Synbiotic | 98.3 ± 4.1 | 92.7 ± 8.2 | 94.1 ± 8.4 | 92.7 ± 11.1 |
| Vomiting (%) | ||||
| − Placebo | 0 ± 0 | 26.8 ± 22.2 | 14.3 ± 20.2 | 9.0 ± 17.0 |
| − Synbiotic | 1.8 ± 5.1 | 21.5 ± 15.3 | 9.0 ± 15.2 | 1.8 ± 5.1 |
| Fecal score | ||||
| − Placebo | 2.6 ± 0.8 | 3.2 ± 0.8 | 3.5 ± 0.8 | 3.7 ± 1.3 |
| − Synbiotic | 2.7 ± 0.8 | 3.7 ± 0.9 | 3.8 ± 0.7 | 4.2 ± 0.4 |
Fecal microbiome
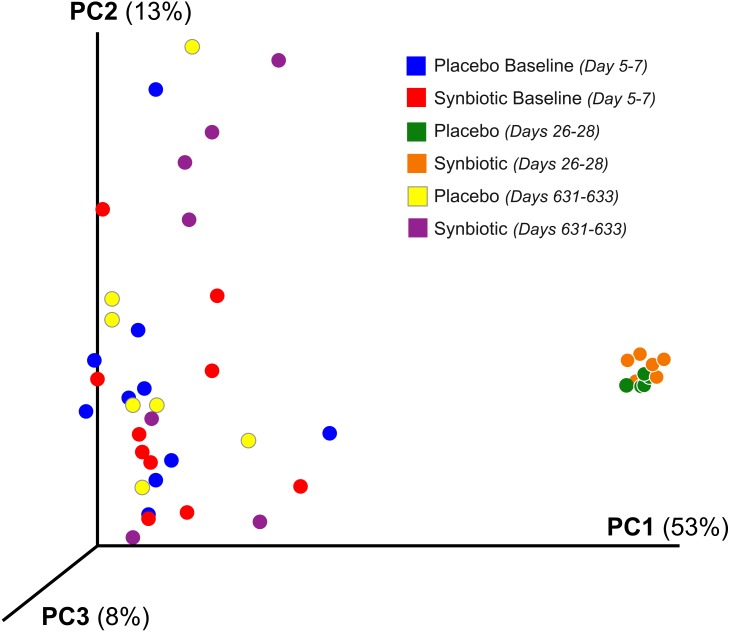
Cats in both treatment groups had marked changes in their fecal microbiome over time (Figs. 2– 3). Alpha diversity was significantly lower (P < 0.01) on days 26–28, based on the Shannon index, goods coverage, and Chao1 metric (Table 3). There was no significant association between treatment group and changes in Shannon index, goods coverage, or the Chao1 metric. Similarly, beta diversity was significantly altered on days 26–28 compared to baseline and days 631–633 in each treatment group (P < 0.01; R = 0.73–0.76) with no significant differences between treatment groups (Fig. 2). The dysbiosis index was significantly higher on days 26–28 than at baseline or on days 631–633 in each treatment group (Table 3), and a significant association was found between treatment group and dysbiosis index (Table 3, F-value 13.6, P < 0.01).
Figure 2. Principal Coordinate Analysis (PCoA) of unweighted UniFrac distances of 16S rRNA genes.
Gene sequences were determined using fecal samples collected at baseline (days 5–7), at the conclusion of antibiotic administration (days 26–28), and after a 603 day washout (days 631–633) from 16 healthy cats, eight per group, + that received 150 mg clindamycin with either a placebo or synbiotic PO once daily for 21 days. +Feces from one cat (synbiotic group) unavailable on days 631–633.
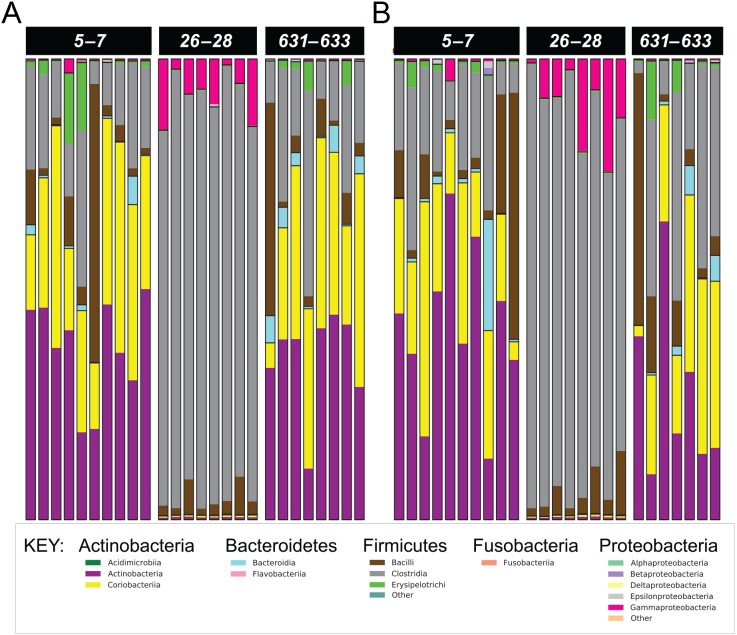
Figure 3. Phylum- and class-level composition of fecal microbiota obtained from feline fecal samples.
(A) Placebo. (B) Synbiotic. Samples were collected at baseline (days 5–7), at the conclusion of antibiotic administration (days 26–28), and after a 603 day washout (days 631–633) from 16 healthy cats, eight per group, + that received 150 mg clindamycin with either a placebo or synbiotic PO once daily for 21 days. +Feces from one cat (synbiotic group) unavailable on days 631–633. Legend for all detected classes is shown, grouped by phylum.
Table 3. Alpha diversity and dysbiosis index results for cats that received clindamycin and placebo or synbiotic.
Median (range) results for feces collected at baseline (days 5–7), at the conclusion of antibiotic administration (days 26–28), and after a 603 day washout (days 631–633) from 16 healthy cats, eight per group, + that received 150 mg clindamycin with either a placebo or synbiotic PO once daily for 21 days. + Feces from one cat (synbiotic group) unavailable at the days 631–633 timepoint. Cells that do not share a common superscript letter differed significantly (P < 0.05) based on post-hoc analysis.
| Baseline | Days 26–28 | Days 631–633 | P-value | ||||
|---|---|---|---|---|---|---|---|
| Placebo | Synbiotic | Placebo | Synbiotic | Placebo | Synbiotic | ||
| Shannon index | 3.9a (3.3–5.3) |
4.2a (3.5–4.8) |
2.5b (1.8–3.0) |
2.8b (2.1–3.1) |
4.4a (3.6–5.2) |
4.9a (3.0–5.6) |
<0.001 |
| Goods coverage | 0.9859a (0.9794–0.9919) |
0.9857a (0.9819–0.9920) |
0.9906b (0.9870–0.9942) |
0.9907b (0.9875–0.9927) |
0.9858a (0.9809–0.9909) |
0.9866a (0.9815–0.9894) |
<0.001 |
| Chao1 metric | 2,085a (1,126–3,158) |
1,960a (1,044–2,930) |
1,419b (830–2,026) |
1,408b (1,043–2,062) |
2,119a (1,272–2,945) |
1,976a (1,349–2,622) |
<0.001 |
| Dysbiosis index | −2.9c (−5.5 to −1.6) |
−3.0c (−5.2 to −0.2) |
1.6b (0.4 to 3.3) |
4.7a (1.1 to 5.4) |
−2.6c (−5.1 to −1.0) |
−4.4c (−5.2 to −0.4) |
<0.001 |
Five phyla were identified based on sequencing analysis (mean baseline prevalences): Actinobacteria (67.95%), Firmicutes (30.39%), Proteobacteria (1.03%), Bacteroidetes (0.73%), and Fusobacteria (0.02%), with marked and significant differences over time in relative abundances of all phyla except Fusobacteria (Fig. 3, Table 4). There was no significant effect of treatment group on changes in relative bacterial abundances at any timepoint, and there was no significant association between age or sex and changes in any of the analyzed fecal microbiome parameters.
Table 4. Median percent abundance (range) for taxa with significantly different relative abundances over time.
Median (range) percent relative abundances of different taxa in feces were collected at baseline (days 5–7), at the conclusion of antibiotic administration (days 26–28), and after a 603 day washout (days 631–633) from 16 healthy cats, eight per group, + that received 150 mg clindamycin with either a placebo or synbiotic PO once daily for 21 days. + Feces from one cat (synbiotic group) unavailable at the days 631–633 timepoint.
| Baseline | Days 26–28 | Days 631–633 | Fdr-adjusted P-value* | ||||
|---|---|---|---|---|---|---|---|
| Placebo | Synbiotic | Placebo | Synbiotic | Placebo | Synbiotic | ||
| Actinobacteria | 68.02a (34.02–87.12) |
71.32a (55.95–83.97) |
0.96b (0.82–1.07) |
0.92b (0.69–1.20) |
69.43a (38.38–82.91) |
51.75a (31.39–90.03) |
<0.001 |
| •Actinobacteria | 68.02a (34.02–87.12) |
71.32a (55.95–83.97) |
0.96b (0.82–1.07) |
0.92b (0.69–1.20) |
69.43a (38.38–82.91) |
51.75a (31.39–90.03) |
<0.001 |
| −Actinomycetales | 0.04a (0.00–0.07) |
0.04a (0.02–0.06) |
0.00b (0.00–0.01) |
0.00b (0.00–0.01) |
0.03a (0.01–0.13) |
0.03a (0.02–0.16) |
<0.001 |
| o Actinomycetaceae | 0.03a (0.00–0.06) |
0.04a (0.01–0.06) |
0.00b (0.00–0.01) |
0.00b (0.00–0.01) |
0.03a (0.01–0.13) |
0.03a (0.01–0.14) |
<0.001 |
| ■Actinomyces | 0.033a (0.000–0.060) |
0.037a (0.009–0.060) |
0.001b (0.000–0.006) |
0.003b (0.000–0.006) |
0.026a (0.006–0.126) |
0.034a (0.014–0.143) |
<0.001 |
| −Bifidobacteriales | 39.16a (18.91–46.70) |
46.10a (18.05–70.73) |
0.54b (0.45–0.65) |
0.49b (0.42–0.71) |
39.16c (10.98–44.47) |
18.72c (9.85–64.69) |
<0.001 |
| o Bifidobacteriaceae | 39.16a (18.91–46.70) |
46.10a (18.05–70.73) |
0.54b (0.45–0.65) |
0.49b (0.42–0.71) |
39.16c (10.98–44.47) |
18.72c (9.85–64.69) |
<0.001 |
| ■Bifidobacterium | 39.12a (18.91–46.70) |
46.08a (18.01–70.72) |
0.54b (0.45–0.65) |
0.49b (0.42–0.71) |
39.12c (10.97–44.46) |
18.70c (9.82–64.66) |
<0.001 |
| •Coriobacteriia | 27.29a (14.31–48.21) |
21.64a (13.19–50.92) |
0.42b (0.32–0.48) |
0.40b (0.26–0.53) |
34.95a (5.43–46.25) |
25.28a (2.33–38.40) |
<0.001 |
| −Coriobacteriales | 27.29a (14.31–48.21) |
21.64a (13.19–50.92) |
0.42b (0.32–0.48) |
0.40b (0.26–0.53) |
34.95a (5.43–46.25) |
25.28a (2.33–38.40) |
<0.001 |
| o Coriobacteriaceae | 27.29a (14.31–48.21) |
21.64a (13.19–50.92) |
0.42b (0.32–0.48) |
0.40b (0.26–0.53) |
34.95a (5.43–46.25) |
25.28a (2.33–38.40) |
<0.001 |
| ■Adlercreutzia | 0.024a (0.003–0.051) |
0.014a (0.006–0.037) |
0.003b (0.000–0.003) |
0.000b (0.000–0.003) |
0.059c (0.006–0.094) |
0.031c (0.009–0.217) |
<0.001 |
| ■Collinsella | 9.39a (1.39–25.36) |
8.91a (3.19–21.39) |
0.21b (0.17–0.24) |
0.21b (0.16–0.25) |
8.69a (4.17–33.50) |
16.18a (1.88–20.90) |
<0.001 |
| ■Slackia | 0.13a (0.04–0.21) |
0.11a (0.05–0.27) |
0.01b (0.00–0.02) |
0.01b (0.00–0.01) |
0.17a (0.14–0.30) |
0.19a (0.01–0.35) |
<0.001 |
| Bacteroidetes | 0.54a (0.11–2.17) |
0.80a (0.17–1.67) |
0.09b (0.05–0.15) |
0.10b (0.05–0.13) |
3.35a (0.30–5.91) |
0.82a (0.08–6.37) |
<0.001 |
| •Bacteroidia | 0.54a (0.11–2.17) |
0.80a (0.17–1.67) |
0.09b (0.05–0.15) |
0.10b (0.05–0.13) |
3.35a (0.30–5.91) |
0.81a (0.08–6.37) |
<0.001 |
| −Bacteroidales | 0.54a (0.11–2.17) |
0.80a (0.17–1.67) |
0.09b (0.05–0.15) |
0.10b (0.05–0.13) |
3.35a (0.30–5.91) |
0.81a (0.08–6.37) |
<0.001 |
| o Bacteroidaceae | 0.14a (0.07–1.67) |
0.24a (0.10–1.13) |
0.07b (0.04–0.13) |
0.09b (0.04–0.11) |
3.08c (0.21–5.85) |
0.53c (0.07–6.03) |
<0.001 |
| ■Bacteroides | 0.14a (0.07–1.67) |
0.24a (0.10–1.13) |
0.07b (0.04–0.13) |
0.09b (0.04–0.11) |
3.08c (0.21–5.85) |
0.53c (0.07–6.03) |
<0.001 |
| o Porphyromonadaceae | 0.007a (0.003–0.349) |
0.013a (0.006–0.037) |
0.003b (0.003–0.011) |
0.006b (0.003–0.017) |
0.071c (0.014–0.514) |
0.063c (0.003–0.631) |
<0.001 |
| ■Parabacteroides | 0.007a (0.003–0.349) |
0.013a (0.006–0.037) |
0.003b (0.003–0.011) |
0.006b (0.003–0.017) |
0.071c (0.014–0.514) |
0.063c (0.003–0.631) |
<0.001 |
| o Prevotellaceae | 0.19a (0.01–0.63) |
0.36a (0.02–0.79) |
0.009b (0.003–0.014) |
0.006b (0.000–0.011) |
0.10a (0.01–0.21) |
0.14a (0.01–0.28) |
<0.001 |
| ■Prevotella | 0.194a (0.011–0.634) |
0.363a (0.020–0.794) |
0.009b (0.003–0.014) |
0.006b (0.000–0.011) |
0.100a (0.014–0.206) |
0.137a (0.006–0.277) |
<0.001 |
| Firmicutes | 30.29a (11.58–65.48) |
27.64a (10.37–42.67) |
91.84b (83.66–97.50) |
90.75b (74.36–98.31) |
26.21a (13.98–55.26) |
47.08a (8.84–67.65) |
<0.001 |
| •Clostridia | 12.69a (4.63–33.88) |
18.83a (6.76–35.33) |
85.49b (81.13–95.00) |
82.78b (70.84–96.36) |
15.95a (7.95–44.59) |
36.75a (2.44–45.25) |
<0.001 |
| −Clostridiales | 12.69a (4.63–33.88) |
18.83a (6.76–35.33) |
85.49b (81.13–95.00) |
82.78b (70.84–96.36) |
15.95a (7.95–44.59) |
36.75a (2.44–45.25) |
<0.001 |
| o Clostridiales; Other | 0.25a (0.10–1.14) |
0.31a (0.05–0.97) |
0.06b (0.03–0.13) |
0.05b (0.03–0.13) |
0.79a (0.09–2.01) |
0.54a (0.01–2.47) |
<0.001 |
| ■ Other | 0.25a (0.10–1.14) |
0.31a (0.05–0.97) |
0.06b (0.03–0.13) |
0.05b (0.03–0.13) |
0.79a (0.09–2.01) |
0.54a (0.01–2.47) |
<0.001 |
| o Clostridiaceae | 1.97a (1.32–6.98) |
2.28a (1.56–11.82) |
82.66b (78.23–92.91) |
80.16b (68.32–94.47) |
3.56a (1.23–22.91) |
5.99a (1.12–24.29) |
<0.001 |
| ■ Other | 0.77a (0.47–2.22) |
0.75a (0.47–4.83) |
66.32b (60.30–77.61) |
57.64b (54.96–72.51) |
0.67a (0.48–2.33) |
0.82a (0.44–5.27) |
<0.001 |
| ■ ___ | 0.83a (0.57–5.43) |
1.40a (0.82–10.25) |
8.52b (3.79–12.68) |
7.67b (3.53–13.62) |
2.71c (0.65–18.51) |
4.76c (0.47–21.95) |
<0.001 |
| ■Clostridium | 0.16a (0.11–0.25) |
0.16a (0.08–0.47) |
9.97b (8.78–11.13) |
9.20b (7.90–11.19) |
0.15a (0.09–1.63) |
0.19a (0.11–0.55) |
<0.001 |
| ■ SMB53 | 0.020a (0.009–0.151) |
0.026a (0.014–0.389) |
0.011b (0.009–0.026) |
0.016b (0.011–0.077) |
0.089c (0.014–0.740) |
0.197c (0.009–0.817) |
<0.001 |
| o Lachnospiraceae | 6.71a (1.66–16.53) |
9.12a (1.05–14.87) |
1.40b (1.14–1.79) |
1.46b (1.06–1.74) |
7.92a (2.47–16.14) |
14.56a (0.75–20.25) |
<0.001 |
| ■ Other | 0.33a (0.05–0.73) |
0.68a (0.07–1.50) |
0.06b (0.04–0.11) |
0.06b (0.03–0.09) |
0.77a (0.27–1.28) |
0.78a (0.05–1.97) |
<0.001 |
| ■Blautia | 4.75a (1.16–10.51) |
6.16a (0.42–8.29) |
0.58b (0.48–0.69) |
0.60b (0.46–0.73) |
4.49a (1.39–12.61) |
10.91a (0.36–14.02) |
0.002 |
| ■Coprococcus | 0.016a (0.011–0.066) |
0.027a (0.011–0.091) |
0.014b (0.009–0.031) |
0.013b (0.003–0.023) |
0.050c (0.014–0.117) |
0.086c (0.000–0.203) |
0.002 |
| ■Dorea | 0.52a (0.20–1.40) |
0.54a (0.16–0.96) |
0.35a (0.30–0.47) |
0.33a (0.24–0.44) |
0.69b (0.31–1.41) |
1.01b (0.18–2.23) |
0.014 |
| ■Roseburia | 0.011a (0.003–0.151) |
0.013a (0.003–0.040) |
0.007a (0.000–0.020) |
0.009a (0.000–0.023) |
0.031b (0.026–0.160) |
0.040b (0.003–0.280) |
<0.001 |
| ■ [Ruminococcus] | 0.92a (0.17–5.49) |
1.14a (0.24–3.84) |
0.25b (0.20–0.38) |
0.28b (0.18–0.35) |
0.74a (0.21–1.20) |
1.19a (0.11–2.05) |
0.002 |
| o Peptococcaceae | 0.15a (0.06–0.40) |
0.20a (0.03–0.71) |
0.06b (0.02–0.09) |
0.06b (0.03–0.09) |
0.23a (0.04–1.68) |
0.34a (0.01–1.67) |
0.015 |
| ■Peptococcus | 0.15a (0.06–0.40) |
0.20a (0.03–0.71) |
0.06b (0.02–0.09) |
0.06b (0.03–0.09) |
0.23a (0.04–1.68) |
0.34a (0.01–1.67) |
0.014 |
| o Ruminococcaceae | 1.22a (0.25–3.55) |
1.97a (0.14–9.85) |
0.25b (0.18–0.51) |
0.22b (0.17–0.38) |
0.82a (0.16–4.79) |
0.58a (0.11–2.54) |
<0.001 |
| ■Faecalibacterium | 0.010a (0.000–0.146) |
0.031a (0.003–0.077) |
0.003b (0.000–0.009) |
0.001b (0.000–0.011) |
0.006a (0.000–0.063) |
0.011a (0.000–0.091) |
0.016 |
| ■Oscillospira | 0.029a (0.009–0.163) |
0.033a (0.011–0.057) |
0.127b (0.066–0.320) |
0.100b (0.023–0.174) |
0.083b (0.026–0.434) |
0.160b (0.006–0.543) |
<0.001 |
| ■Ruminococcus | 0.017a (0.006–0.031) |
0.017a (0.006–0.040) |
0.009b (0.006–0.017) |
0.010b (0.003–0.014) |
0.034c (0.011–0.160) |
0.037c (0.003–0.054) |
0.002 |
| o Veillonellaceae | 1.03a (0.13–4.81) |
0.97a (0.54–4.88) |
0.07b (0.03–0.12) |
0.08b (0.03–0.16) |
0.26c (0.08–2.63) |
0.17c (0.08–3.91) |
<0.001 |
| ■Megamonas | 0.11a (0.01–0.22) |
0.21a (0.02–2.38) |
0.02b (0.00–0.05) |
0.02b (0.01–0.06) |
0.04a (0.02–0.35) |
0.05a (0.00–0.28) |
0.009 |
| ■Megasphaera | 0.86a (0.08–4.68) |
0.72a (0.08–2.42) |
0.04b (0.02–0.07) |
0.05b (0.02–0.08) |
0.23c (0.05–2.20) |
0.09c (0.04–3.58) |
<0.001 |
| ■Phascolarctobacterium | 0.009a (0.003–0.029) |
0.009a (0.000–0.069) |
0.000b (0.000–0.006) |
0.006b (0.000–0.017) |
0.011a (0.003–0.063) |
0.003a (0.000–0.046) |
0.004 |
| •Erysipelotrichi | 0.81a (0.21–15.32) |
1.05a (0.18–5.59) |
0.28b (0.21–0.36) |
0.30b (0.22–0.37) |
0.89a (0.21–6.46) |
0.78a (0.24–12.78) |
0.008 |
| −Erysipelotrichiales | 0.81a (0.21–15.32) |
1.05a (0.18–5.59) |
0.28b (0.21–0.36) |
0.30b (0.22–0.37) |
0.89a (0.21–6.46) |
0.78a (0.24–12.78) |
0.005 |
| o Erysipelotrichaceae | 0.81a (0.21–15.32) |
1.05a (0.18–5.59) |
0.28b (0.21–0.36) |
0.30b (0.22–0.37) |
0.89a (0.21–6.46) |
0.78a (0.24–12.78) |
0.006 |
| ■Bulleidia | 0.019a (0.006–0.194) |
0.006a (0.000–0.123) |
0.003b (0.000–0.009) |
0.004b (0.000–0.011) |
0.016a (0.000–0.431) |
0.009a (0.003–0.014) |
0.015 |
| ■[Eubacterium] | 0.26a (0.08–6.60) |
0.22a (0.04–1.17) |
0.07b (0.05–0.12) |
0.08b (0.03–0.13) |
0.27a (0.08–2.94) |
0.33a (0.10–6.63) |
<0.001 |
| Proteobacteria | 0.39a (0.24–3.03) |
0.48a (0.29–4.74) |
7.09b (1.28–15.46) |
8.34b (0.85–24.53) |
0.43a (0.28–0.57) |
0.43a (0.21–0.89) |
<0.001 |
| •Gammaproteobacteria | 0.27a (0.16–2.98) |
0.34a (0.18–4.63) |
7.04b (1.23–15.32) |
8.30b (0.83–24.39) |
0.25c (0.14–0.37) |
0.31c (0.18–0.37) |
<0.001 |
| −Aeromonadales | 0.01a (0.00–0.07) |
0.02a (0.01–0.04) |
0.03b (0.02–0.07) |
0.03b (0.01–0.05) |
0.01a (0.00–0.03) |
0.01a (0.01–0.04) |
0.010 |
| o Succinivibrionaceae | 0.009a (0.003–0.069) |
0.021a (0.006–0.043) |
0.033b (0.017–0.071) |
0.030b (0.014–0.051) |
0.014a (0.003–0.029) |
0.014a (0.006–0.040) |
0.013 |
| ■Anaerobiospirillumi | 0.007a (0.003–0.060) |
0.017a (0.006–0.029) |
0.031b (0.017–0.071) |
0.030b (0.014–0.051) |
0.014a (0.003–0.023) |
0.011a (0.006–0.037) |
0.002 |
| −Enterobacteriales | 0.26a (0.15–2.92) |
0.32a (0.14–4.62) |
6.99b (1.19–15.30) |
8.25b (0.81–24.34) |
0.23c (0.13–0.34) |
0.28c (0.17–0.33) |
<0.001 |
| o Enterobacteriaceae | 0.26a (0.15–2.92) |
0.32a (0.14–4.62) |
6.99b (1.19–15.30) |
8.25b (0.81–24.34) |
0.23c (0.13–0.34) |
0.28c (0.17–0.33) |
<0.001 |
Notes.
P-values adjusted based on the Benjamini and Hochberg False discovery rate (fdr).
Relative abundances that do not share a common superscript letter differed significantly (fdr-adjusted P < 0.05) based on post-hoc analysis.
Metabolome
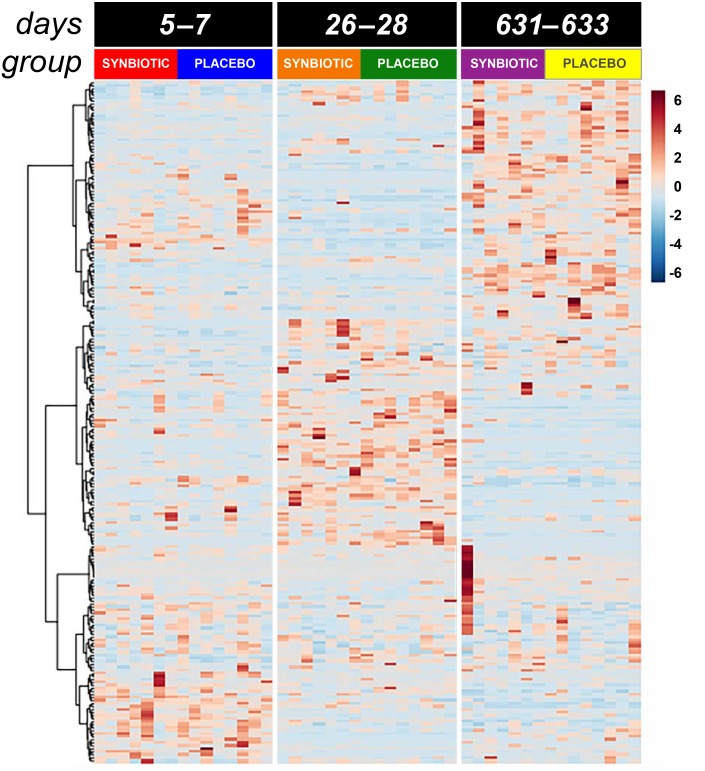
Untargeted fecal metabolomics profiles differed markedly over time for cats in both treatment groups (Figs. 4– 5), with a variety of patterns of changes found. Fecal profiles for 178 of 252 assayed compounds (71%) differed significantly over time (see Additional file 1). Of those that differed over time, 47 of 178 (26.4%) metabolites had equivalent profiles at baseline and days 631–633. Partial return to baseline profiles were identified for an additional 14 metabolites (7.9%), and 21 metabolites (11.8%) had altered profiles on days 26–28 compared to baseline with no significant changes between days 26–28 and days 631–633. Changes in profiles for the remaining 96 metabolites (53.9%) were more complex. Profiles for 33 compounds (18.5%) differed significantly on days 631–633 from both baseline and days 26–28 with no difference between baseline and days 26–28, and profiles for 14 metabolites (7.9%) were significantly higher or lower at days 26–28 compared to baseline with further significant changes in the same direction by days 631–633. In contrast, profiles for 30 metabolites (16.9%) were significantly higher at days 26–28 than baseline but also significantly lower at days 631–633 than baseline; profiles for another 14 metabolites (7.9%) were decreased on days 26–28 with much higher profiles on days 631–633 than baseline. Finally, 5 metabolites had individual patterns of change.
Figure 4. Dual hierarchical dendrogram of metabolites, clustered by pathway, that differed significantly over time in feline fecal samples.
Samples were collected at baseline (days 5–7), at the conclusion of antibiotic administration (days 26–28), and after a 603 day washout (days 631–633) from 16 healthy cats, 8 per group, + that received 150 mg clindamycin with either a placebo or synbiotic PO once daily for 21 days. + Feces from one cat (synbiotic group) unavailable on days 631–633.
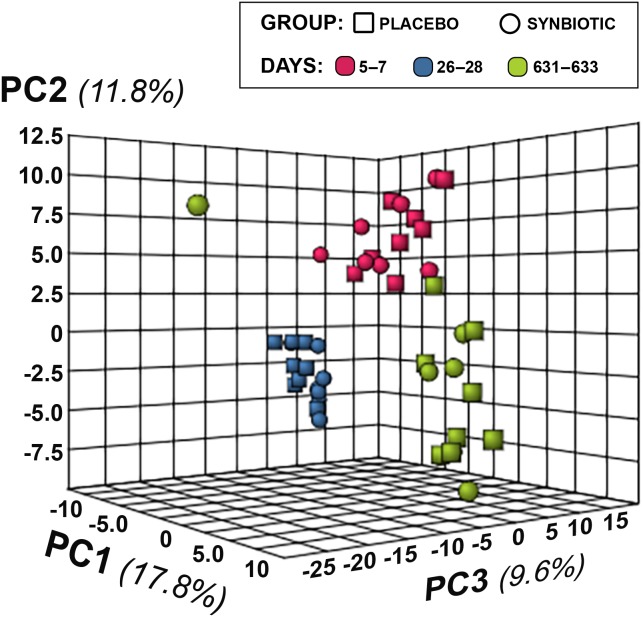
Figure 5. Principal Component Analysis (PCA) of metabolic pathway analyses from feline fecal samples.
Samples were collected at baseline (days 5–7), at the conclusion of antibiotic administration (days 26–28), and after a 603 day washout (days 631–633) from 16 healthy cats, eight per group, + that received 150 mg clindamycin with either a placebo or synbiotic PO once daily for 21 days. +Feces from one cat (synbiotic group) unavailable on days 631–633.
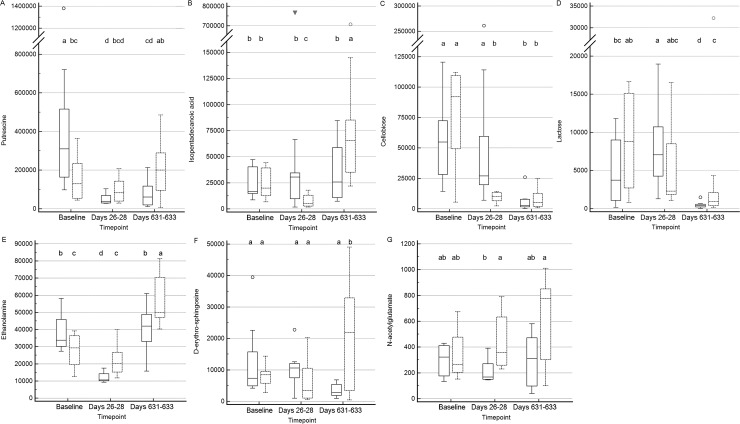
Significant differences (fdr adjusted P < 0.05) were noted between treatment groups for seven metabolite profiles (Fig. 6). Significant group by time interactions were found for fecal profiles of putrescine, isopentadecanoic acid, cellobiose, ethanolamine, lactose, and D-erythro-sphingosine, while fecal profiles for N-acetylglutamate differed significantly by treatment group alone.
Figure 6. Box and whisker plots of fecal metabolite profiles for seven fecal metabolites that differed significantly (fdr adjusted P < 0.05) between treatment groups and over time.
(A) Putrescine. (B) Isopentadecanoic acid. (C) Cellobiose. (D) Lactose. (E) Ethanolamine. (F) D-erythro-sphingosine. (G) N-acetylglutamate. Medians, interquartile ranges, and minimum and maximum values are presented for cats in the placebo (boxes with solid borders) and synbiotic (boxes with dashed borders) treatment groups. Open circles and closed triangles denote outlier values. Boxes that do not share a letter differed significantly (fdr-adjusted P < 0.05) based on post-hoc analysis. Fecal samples were collected at baseline (days 5–7), at the conclusion of antibiotic administration (days 26–28), and after a 603 day washout (days 631–633 from 16 healthy cats, 8 per group, + that received 150 mg clindamycin with either a placebo or synbiotic PO once daily for 21 days. +Feces from one cat (synbiotic group) unavailable on days 631–633. Significance was set as P < 0.05, with P- values adjusted based on the Benjamini and Hochberg False discovery rate (fdr).
With regard to metabolites of known biological importance, there were significant changes in profiles of some short-chain fatty acid (SCFA) synthesis metabolites, bile acids, tryptophan degradation pathway metabolites, sphingolipids, polyamines, and benzoic and cinnaminic acids over time (Table 5). Pathway analysis revealed significant changes (fdr adjusted P < 0.05) over time for 55 metabolic pathways; 47 of which had impact factors >0 (Table 6). The majority of pathways affected were related to SCFA, amino acid, sugar, and nucleotide synthesis and degradation. Amino acid pathways that were affected included those related to tryptophan degradation. Other pathways significantly affected included those related to linoleic acid, sphingolipid, and glycerolipid metabolism.
Table 5. Metabolites of known biological import with profiles that significantly differed over time.
Median (range) peak height of metabolites in feces collected at baseline (days 5–7), at the conclusion of antibiotic administration (days 26–28), and after a 603 day washout (days 631–633) from 16 healthy cats, eight per group, + that received 150 mg clindamycin with either a placebo or synbiotic PO once daily for 21 days. + Feces from one cat (synbiotic group) unavailable at days 631–633.
| Baseline | Days 26–28 | Days 631–633 | Fdr-adjusted P-value* | ||||
|---|---|---|---|---|---|---|---|
| Placebo | Synbiotic | Placebo | Synbiotic | Placebo | Synbiotic | ||
| Short chain fatty acid metabolites | |||||||
| 3,4-dihydroxyphenylacetic acid | 589a (317–1,201) |
521a (221–1,577) |
741a (263–1,130) |
594a (126–1,522) |
197b (66–1,436) |
199b (63–643) |
<0.001 |
| 3-hydroxyphenylacetic acid | 3,350a (582–12,188) |
2,860a (55–11,401) |
65b (36–94) |
57b (26–239) |
554c (77–3,578) |
275c (124–758) |
<0.001 |
| 4-hydroxyphenylacetic acid | 24,894a (10,957–34,200) |
24,248a (12,980–73,736) |
693b (473–6,935) |
3,268b (378–139,343) |
48,775a (10,270–133,645) |
47,322a (837–128,116) |
<0.001 |
| 2,3-dihydroxybutanoic acid | 167a (119–347) |
193a (67–240) |
83b (18–157) |
108b (25–268) |
216a (97–356) |
128a (63–376) |
<0.001 |
| 2-hydroxybutanoic acid | 12,514a (3,401–161,892) |
10,110a (626–61,119) |
38,839b (26,636–58,406) |
38,680b (18,670–76,553) |
3,846c (1,170–13,209) |
3,082c (623–13,862) |
<0.001 |
| 4-aminobutyric acid | 2,002a (1,665–12,898) |
2,573a (1,189–11,658) |
112,677b (72,970–190,000) |
156,808b (56,870–393,600) |
26,744c (3,966–89,927) |
20,753c (437–60,568) |
<0.001 |
| 2,4-diaminobutyric acid | 1,758a (568–3,102) |
1,119a (184–3,865) |
210b (140–350) |
254b (109–590) |
3,095c (1,652–8,818) |
3,017c (765–6,398) |
<0.001 |
| 3-phenyllactic acid | 5,539a (781–31,484) |
5,796a (438–32,942) |
9,109b (7,089–11,018) |
12,813b (7,617–21,692) |
6,537a (1,217–19,572) |
9,052a (331–16,813) |
<0.001 |
| Lactic acid | 152,579a (24,216–1,034,220) |
120,122a (10,433–1,403,821) |
392,894b (269,519–1,091,298) |
308,949b (129,795–638,477) |
27,338c (11,859–149,087) |
35,473c (6,439–111,809) |
<0.001 |
| P-hydroxylphenyl-lactic acid | 3,698a (1,153–6,357) |
4,503a (581–12,311) |
3,590a (2,472–6,688) |
4,760a (2,300–5,993) |
1,654b (478–5,019) |
1,431b (405–2,022) |
<0.001 |
| Propane-1-3-diol | 1,108a (426–15,239) |
1,900a (272–5,779) |
12,212b (3,684–26,363) |
12,461b (4,117–20,201) |
359c (174–1,203) |
341c (99–502) |
<0.001 |
| 3,3-hydroxyphenyl propionic acid | 76,962a (18,783–212,470) |
45,560a (865–240,661) |
789b (88–1,348) |
648b (188–1,431) |
77,032a (666–167,768) |
27,581a (140–134,885) |
<0.001 |
| 3-(4-hydroxyphenyl)-propionic acid | 65,664a (28,474–90,967) |
56,101a (4,122–141,022) |
4,255b (350–16,751) |
6,447b (415–16,156) |
28,844c (4,747–68,675) |
14,394c (12,923–79,425) |
<0.001 |
| Pyruvic acid | 7,118a (1,844–19,684) |
7,900a (3,534–12,585) |
7,744a (3,418–20,148) |
9,650a (1,509–48,246) |
915b (456–3,736) |
1,588b (489–2,230) |
<0.001 |
| Succinic acid | 236,185a (14,243–708,079) |
88,426a (882–1,122,477) |
41,400a (25,669–96,357) |
48,077a (7,555–107,389) |
8,572b (3,020–65,677) |
9,082b (1,337–65,270) |
<0.001 |
| Bile acids | |||||||
| Cholic acid | 109,466a (1,898–200,811) |
59,483a (423–220,771) |
11,303b (113–76,704) |
27,391b (286–89,000) |
9,511b (240–77,925) |
8,907b (3,191–95,296) |
0.04 |
| Deoxycholic acid | 3,295a (77–28,703) |
1,843a (109–43,646) |
136b (45–5,010) |
352b (27–3,531) |
11,544a (150–111,319) |
8,762a (687–309,022) |
0.01 |
| Tryptophan metabolites | |||||||
| Indole-3-acetate | 13,713a (7,780–94,057) |
8,832a (3,110–125,618) |
841b (541–1,795) |
1,051b (448–11,089) |
15,203a (6,967–22,835) |
27,026c (17,204–168,850) |
<0.001 |
| Indole-3-lactate | 80,846a (34,669–127,305) |
62,189a (7,536–128,029) |
6,751b (2,893–11,123) |
7,133b (4,437–22,490) |
15,933b (773–44,962) |
5,311b (836–54,443) |
<0.001 |
| Tryptophan | 190,390a (132,057–340,750) |
173,956a (106,947–276,525) |
120,418b (95,182–158,327) |
153,738b (19,076–294,367) |
20,219c (3,724–138,699) |
35,369c (5,707–174,485) |
<0.001 |
| Sphingolipid metabolites | |||||||
| Cellobiose | 54,905a (14,235–120,530) |
92,176a (5,399–111,969) |
28,177ab (6,974–261,178) |
10,182b (2,284–14,269) |
2,513b (249–26,017) |
5,248b (753–24,528) |
<0.001 |
| D-erythro-sphingosine | 7,341a (4,159–39,450) |
8,433a (2,795–14,346) |
10,575a (1,003–22,798) |
3,460a (620–20,251) |
2,778b (869–6,781) |
21,981a (438–49,033) |
<0.001 |
| Isopentadecanoic acid | 16,537b (8,419–47,233) |
19,852a (6,576–44,308) |
32,099b (1,673–766,411) |
5,119b (1,627–17,766) |
25,695b (7,307–84,652) |
65,848c (21,866–707,919) |
<0.001 |
| Polyamines | |||||||
| Putrescine | 325,567a (97,835–1,380,413) |
129,926a (44,868–363,883) |
38,927b (27,370–102,805) |
83,581a (28,669–207,557) |
60,980a (11,066–212,790) |
201,011a (6,192–485,041) |
0.01 |
| Antioxidants / antimicrobials | |||||||
| 3,4-dihydroxyhydrocinnamic acid | 20,308a (11,158–79,529) | 19,951a (9,793–36,089) |
6,608b (3,079–14,086) |
7,699b (4,398–16,818) |
28,082a (1,849–40,617) |
26,438a (1,485–61,532) |
<0.001 |
| 3,4-dihydroxycinnaminic acid | 4,356a (1,269–10,007) |
6,229a (1,990–25,507) |
2,543a (1,178–16,989) |
2,955a (1,481–18,549) |
1,468b (281–6,545) |
760b (161–4,120) |
<0.001 |
| 3,4-dihydroxybenzoic acid | 4,141a (1,918–12,984) |
4,062a (2,054–13,140) |
2,114b (1,533–3,328) |
2,489b (1,694–4,578) |
13,395c (2,783–26,592) |
6,790c (518–26,569) |
<0.001 |
| 4-hydroxybenzoate | 6,538a (3,972–27,743) |
11,523a (4,231–17,030) |
1,035b (665–1,411) |
1,201b (619–1,940) |
16,773c (8,232–23,719) |
16,643c (420–49,060) |
<0.001 |
Notes.
P-values adjusted based on the Benjamini and Hochberg False discovery rate (fdr).
Profiles that do not share a common superscript letter differed significantly (fdr-adjusted P < 0.05) based on post-hoc analysis.
Table 6. Metabolic pathways that differed significantly over time.
Pathways that differed significantly over time by group based on fecal metabolite profiles in feces collected at baseline (days 5–7), at the conclusion of antibiotic administration (days 26–28), and after a 603 day washout (days 631–633) from 16 healthy cats, eight per group, + that received 150 mg clindamycin with either a placebo or synbiotic PO once daily for 21 days. +Feces from one cat (synbiotic group) unavailable at the days 631–633 timepoint.
| Total Cmpds | Hits | fdr-adjusted P-value | Pathway impact value | ||
|---|---|---|---|---|---|
| 1 | Linoleic acid metabolism | 15 | 1 | 0.018 | 0.66 |
| 2a | Alanine, aspartate and glutamate metabolism | 24 | 9 | 1.54E–10 | 0.59 |
| 3b | Galactose metabolism | 41 | 16 | 2.00E–08 | 0.58 |
| 4a | Glycine, serine and threonine metabolism | 48 | 9 | 4.69E–09 | 0.54 |
| 5a | Arginine and proline metabolism | 77 | 16 | 1.54E–10 | 0.48 |
| 6a | Taurine and hypotaurine metabolism | 20 | 4 | 1.37E–08 | 0.38 |
| 7b | Starch and sucrose metabolism | 50 | 8 | 2.87E–08 | 0.36 |
| 8a | beta-Alanine metabolism | 28 | 6 | 7.25E–09 | 0.34 |
| 9 | Pyruvate metabolism | 32 | 2 | 3.82E–06 | 0.32 |
| 10a | Phenylalanine metabolism | 45 | 13 | 1.18E–08 | 0.27 |
| 11c | Pantothenate and CoA biosynthesis | 27 | 6 | 2.77E–09 | 0.25 |
| 12a | Cysteine and methionine metabolism | 56 | 8 | 4.43E–09 | 0.23 |
| 13 | Nicotinate and nicotinamide metabolism | 44 | 4 | 1.43E–08 | 0.23 |
| 14a | Lysine degradation | 47 | 4 | 1.50E–02 | 0.23 |
| 15d | Pyrimidine metabolism | 60 | 9 | 6.20E–08 | 0.22 |
| 16 | Arachidonic acid metabolism | 62 | 1 | 1.01E–04 | 0.22 |
| 17 | Glycerolipid metabolism | 32 | 5 | 2.80E–08 | 0.22 |
| 18 | Histidine metabolism | 44 | 3 | 1.43E–08 | 0.21 |
| 19a,d | Amino sugar and nucleotide sugar metabolism | 88 | 9 | 1.37E–08 | 0.21 |
| 20a | Tyrosine metabolism | 76 | 10 | 1.96E–09 | 0.20 |
| 21 | Aminoacyl-tRNA biosynthesis | 75 | 18 | 2.84E–10 | 0.17 |
| 22c | Butanoate metabolism | 40 | 7 | 9.61E–11 | 0.17 |
| 23a | Tryptophan metabolism | 79 | 2 | 2.77E–09 | 0.16 |
| 24a | D-Glutamine and D-glutamate metabolism | 11 | 2 | 3.82E–06 | 0.14 |
| 25 | Inositol phosphate metabolism | 39 | 1 | 2.60E–03 | 0.14 |
| 26 | Citrate cycle (TCA cycle) | 20 | 3 | 9.75E–06 | 0.12 |
| 27 | Sulfur metabolism | 18 | 4 | 1.59E–08 | 0.11 |
| 28b | Pentose phosphate pathway | 32 | 5 | 8.38E–12 | 0.11 |
| 29d | Purine metabolism | 92 | 12 | 1.61E–10 | 0.11 |
| 30a | Phenylalanine, tyrosine and tryptophan biosynthesis | 27 | 5 | 4.90E–09 | 0.11 |
| 31a | Lysine biosynthesis | 32 | 2 | 3.07E–03 | 0.10 |
| 32 | Glycolysis or Gluconeogenesis | 31 | 4 | 2.12E–06 | 0.10 |
| 33 | Sphingolipid metabolism | 25 | 3 | 5.39E–04 | 0.09 |
| 34b | Pentose and glucuronate interconversions | 53 | 7 | 7.42E–09 | 0.09 |
| 35 | Glycerophospholipid metabolism | 39 | 2 | 3.38E–07 | 0.09 |
| 36c | Propanoate metabolism | 35 | 6 | 2.23E–06 | 0.09 |
| 37a | Valine, leucine and isoleucine biosynthesis | 27 | 6 | 9.82E–07 | 0.09 |
| 38b | Fructose and mannose metabolism | 48 | 3 | 4.94E–04 | 0.07 |
| 39 | Nitrogen metabolism | 39 | 10 | 2.77E–09 | 0.07 |
| 40a | Valine, leucine and isoleucine degradation | 40 | 4 | 1.84E–04 | 0.06 |
| 41 | Ubiquinone and other terpenoid-quinone biosynthesis | 36 | 4 | 2.84E–10 | 0.05 |
| 42 | Glyoxylate and dicarboxylate metabolism | 50 | 4 | 1.18E–06 | 0.04 |
| 43 | Caffeine metabolism | 21 | 1 | 1.84E–04 | 0.03 |
| 44 | Glutathione metabolism | 38 | 5 | 3.28E–10 | 0.03 |
| 45 | Ascorbate and aldarate metabolism | 45 | 3 | 2.59E–08 | 0.02 |
| 46 | Vitamin B6 metabolism | 32 | 1 | 1.47E–05 | 0.02 |
| 47 | Methane metabolism | 34 | 2 | 1.30E–05 | 0.02 |
Notes.
- Total Cmpds
- total compounds in pathway
amino acid metabolism.
sugar metabolism.
short-chain fatty acid metabolism.
nucleotide metabolism.
Discussion
The incidence of AAGS varies in people based on individual antibiotic agent, prior antibiotic exposure, subject age, and other factors (Hempel et al., 2012; Lenoir-Wijnkoop et al., 2014; McFarland, 2008; Ouwehand et al., 2016). Antibiotics, such as clindamycin, can cause AAGS directly by stimulating the intestinal motilin receptor (Bartlett, 2002). However, antibiotic-induced dysbiosis, with secondary alterations in fecal metabolites and overgrowth of opportunistic pathogens, is believed to be the primary contributor to AAGS (Gustafsson et al., 1998; McFarland, 2008; Varughese, Vakil & Phillips, 2013; Videlock & Cremonini, 2012). Dysbiosis is greatest for broad-spectrum and poorly-absorbed antibiotics, including macrolides, cephalosporins, and fluoroquinolones (Jakobsson et al., 2010; Löfmark et al., 2006; McFarland, 2008; Ouwehand et al., 2016), and alterations to the microbiome can persist at least 4 years after short-term antibiotic therapy (Jakobsson et al., 2010; Löfmark et al., 2006).
Multiple metanalyses support the efficacy of probiotics in preventing AAGS in people (Goldenberg et al., 2015; Lau & Chamberlain, 2016; McFarland, 2016; Ouwehand et al., 2016; Szajewska, Konarska & Kołodziej, 2016; Videlock & Cremonini, 2012). Although some gastrointestinal effects appear to be dose-related (Ouwehand et al., 2016; Pagnini et al., 2010; Szajewska, Konarska & Kołodziej, 2016), effects do not correlate directly with the relative abundances of probiotic bacteria or measurable shifts in OTUs in the fecal microbiome (Biagi et al., 2013; Garcia-Mazcorro et al., 2011; Marshall-Jones et al., 2006; Rossi et al., 2014). The ideal microbial strain(s), dose, and timing relative to antibiotic administration likely vary by antibiotic, underlying disease processes, patient (age, inpatient status, historical antibiotic exposure), and other factors (Lau & Chamberlain, 2016; McFarland, 2009; Szajewska, Konarska & Kołodziej, 2016). Positive effects of probiotics include stimulation of the host’s innate system and epithelial secretion of antimicrobial substances, restoration of intestinal barrier function, alteration of the gastrointestinal metabolome, disruption of biofilms through production of bacteriocins, generation of other antimicrobial agents, and competitive exclusion of pathogens (Ohland & MacNaughton, 2010; Ouwehand et al., 2016; Pagnini et al., 2010; Quigley, 2012; Rossi et al., 2014; Strompfová, Lauková & Ouwehand, 2004).
Although AAGS are a recognized complication of antibiotic therapy in cats (Albarellos & Landoni, 2009; Bureau of Veterinary Drugs, 1995; Hunter et al., 1995), the incidence of AAGS, short- and long-term effects on the microbiome and metabolome, and impact of concurrent synbiotic administration had not been previously characterized. In this study, AAGS developed in 100% of cats treated with high dose clindamycin once daily for 21 days. Vomiting was most severe in the first week of therapy, although it persisted for the duration of treatment in some cats. A temporal association between antibiotic administration and vomiting was not noted, suggesting stimulation of the motilin receptor by clindamycin had minimal impact on the incidence of vomiting. The incidence of vomiting per week was less in cats receiving the synbiotic, but the difference between treatment groups was not statistically significant. Fecal consistency and food intake declined progressively throughout the course of treatment in both treatment groups.
There are a number of potential explanations for the lack of statistically significant differences in AAGS between treatment groups in this study. A high antibiotic dose was used to increase the likelihood of detecting a difference between groups because the incidence of AAGS due to clindamycin was unknown. Ironically, this might have obscured positive results due to the severity of AAGS induced. The synbiotic dose chosen for the study was based on a prior report describing clinical improvement in cats with chronic enteropathy treated with the same synbiotic (Hart et al., 2012), but use of a higher dose might be required to prevent AAGS. We elected to administer the synbiotic concomitantly with clindamycin because compliance with medication regimens is inversely correlated with the number or frequency of treatments (Adams et al., 2005; van den Bemt et al., 2016), primarily due to forgetfulness and/or logistical difficulties of balancing treatment schedules against other daily responsibilities (Col, Fanale & Kronholm, 1990). Although macrolide resistance occurs in a relatively high percentage of Enterococcus faecium and Lactobacillus strains, as well as a Bifidobacterium animalis probiotic (European Food Safety Authority, 2012; Ouwehand et al., 2016; Strompfová, Lauková & Ouwehand, 2004), antibiotic sensitivity of the bacterial strains in the study drug were not determined so neutralization due to concomitant antibiotic administration cannot be ruled out.
To further assess the potential efficacy of synbiotics in preventing AAGS in cats, our group has since performed a follow-up study (Stokes, Price & Whittemore, 2017) using a crossover design with a six-week washout period, a more clinically relevant clindamycin dose (mean 18 mg/kg/d), a higher synbiotic dose (10 billion cfu/d), and delaying administration of the synbiotic for 1 h after antibiotic administration to limit potential synbiotic neutralization. Cats receiving the synbiotic were significantly more likely to complete the initial treatment course, vomited less, and had higher food intake than cats receiving the placebo. Interesting, strong period effects were noted—suggesting ameliorative effects of synbiotics on AAGS may persist for at least 6 weeks after administration. Because the antibiotic and synbiotic doses, as well as timing of synbiotic administration, were all altered in that study, it is not possible to ascertain which factor had the greatest impact on clinical signs.
In addition to causing AAGS, clindamycin administration was associated with marked changes in the fecal microbiome and metabolome in this study. Alterations in alpha diversity (Shannon index, Goods coverage, Chao1 metric), beta diversity, and the dysbiosis index returned to baseline levels by the end of the washout period. The return of diversity at a global level was not due to return to baseline abundances for individual OTUs, however. Instead, changes in relative abundances of many individual OTUs persisted through the terminal sampling timepoint (>600 days after discontinuation of antibiotic therapy) in both treatment groups. Changes included reductions in the abundances of Actinobacteria, including Bifidobacterium, Collinsella, and Slackia; Bacteroidetes, including Bacteroides; many members of the Clostridium clusters IV and XIV, including Lachnospiraceae (Blautia, Coprococcus, and Roseburia), Ruminococcaceae (Faecalibacterium and Ruminococcus), and Erysipelotrichaceae (Bulleidia and [Eubacterium ]); and Veillonellaceae, including Megamonas. Marked increases in abundance were identified for Clostridiaceae, including Clostridium, and Proteobacteria, including Aeromonadales but primarily due to increases in Enterobacteriaceae. Alterations were generally consistent with those found in cats (Ramadan et al., 2014; Suchodolski et al., 2015) and people (Schubert et al., 2014) with dysbiosis due to naturally-occurring diarrhea. Interestingly, decreased relative abundances of Lachnospiraceae and Ruminococcaceae also have been found in cats infected with Giardia and feline immunodeficiency virus (Šlapeta et al., 2015; Weese et al., 2015).
Antibiotic-induced dysbiosis and gastrointestinal signs have been associated with alterations in a variety of metabolite profiles. Consistent with prior work in people (Gustafsson et al., 1998; McFarland, 2008; Woodmansey et al., 2004), we found marked derangements in SCFA profiles and pathways during antibiotic administration, some of which persisted at days 631–633. No difference was found in SCFA profiles between treatment groups. In addition to being an important energy source for colonocytes, SCFAs have anti-inflammatory and immunomodulatory effects, regulate intestinal motility, and maintain intestinal barrier function (Quigley, 2012; Suchodolski, 2016a). Persistent alterations in SCFA profiles could perpetuate ongoing dysbiosis after resolution of the initial insult.
Decreased proportions of secondary bile acids, important for maintaining eubiosis and modulating the immune system, have been found in people and dogs with gastrointestinal disease (Duboc et al., 2013; Honneffer et al., 2015). Similarly, we identified a marked reduction in secondary versus primary bile acids during antibiotic administration, followed by an increase at days 631–633 above baseline. It is possible, though speculative, that the inversion in the ratio of primary to secondary bile acids on days 631–633 reflects a compensatory response to persistent, occult dysbiosis and inflammation. Temporal alterations in bile acid biosynthesis were also noted on pathway analysis but did not achieve significance (fdr adjusted P = 0.06).
Indole profiles, the product of tryptophan degradation by commensal E. coli, have previously been found to be decreased in people and animals with gastrointestinal disease (Suchodolski, 2016a). In this study, indole profiles decreased significantly during antibiotic administration in both treatment groups. Indole profiles failed to return to baseline values by days 631–633 for cats in the placebo group, while they were mixed for cats in the synbiotic group. Direct effects of indole within the gut lumen include decreased production of adherence and virulence factors, motility, biofilm formation, and adherence of pathogenic E. coli (Bansal et al., 2007), while effects on the intestine include strengthening of intestinal tight junctions, increased production of mucin to shield the epithelium from bacterial adherence, and modulation of inflammation (Bansal et al., 2010). Persistent alterations in indole profiles in cats not administered a synbiotic with antibiotics could contribute to a pro-inflammatory state, as well as chronically increased intestinal permeability and risk of bacterial colonization and translocation.
Significant temporal changes also were found in profiles of D-erythro-sphingosine, isopentadecanoic acid, and cellobiose. D-erythro-sphingosine is a sphingolipid, while isopentadecanoic acid and cellobiose are components of sphingolipids and sphingophospholipids (Chao, Khan & Hannun, 1992; Minamino et al., 2003; Vesper et al., 1999). Sphingolipids and sphingophospholipids have antineoplastic properties, stimulate the innate immune system, and increase bacterial clearance (Chao, Khan & Hannun, 1992; Fujiwara et al., 2013; Minamino et al., 2003). Persistent reductions in putrescine profiles in cats in the placebo, but not the synbiotic, group after antibiotic exposure also were noted. Putrescine, along with spermine and spermidine, are polyamines. Polyamine supplementation increases resistance to oxidative stress in mice, while depletion is associated with development of pancreatitis (Minois, Carmona-Gutierrez & Madeo, 2011). Further study will be necessary to determine the clinical relevance of differing sphingolipid and putrescine profiles between treatment groups over time.
Finally, significant associations were found in profiles for cinnaminic acid and benzoic acid compounds. Compounds in both treatment groups had lower results at days 26–28 than baseline. Benzoic acid profiles were higher at days 631–633 than at baseline, while cinnaminic profiles did not return to baseline levels. Both cinnaminic and benzoic acids have been found to have antimicrobial effects, and cinnaminic acids have additional anti-inflammatory properties (Alam et al., 2016; Manuja et al., 2013). The persistent decrease in cinnaminic acid profiles after antibiotic administration is particularly intriguing given work in research models suggesting potential benefits of cinnaminic acids in management of obesity and diabetes mellitus (Alam et al., 2016). Repeated exposure to antibiotics has been associated with increased risks of childhood obesity and diabetes mellitus (Mikkelsen et al., 2016; Scott et al., 2016). It is possible, though certainly speculative at this juncture, that associations between antibiotic exposure and increased risk of obesity and diabetes are at least partially due to cumulative derangements in cinnaminic acid profiles.
One factor that could impact clinical application of these results is the use of healthy research cats. Creation of AAGS in healthy privately-owned animals for antibiotic-related investigations poses concerns regarding compliance with the study protocol, while evaluations in clinically ill animals can be confounded by heterogeneity in underlying disease, the antibiotic required for the primary disease, prior antibiotic exposure, and husbandry factors. The predominant phylum identified at baseline in this study, Actinobacteria, conflicted with that found in recent reports in privately-owned healthy cats, Firmicutes (Bell et al., 2014; Suchodolski et al., 2015; Weese et al., 2015), though they agree with findings of other studies, particularly those using research colony cats (Abecia et al., 2010; Desai et al., 2009; Jia et al., 2011a; Jia et al., 2011b). Although Firmicutes was the predominant phyla in the first 3 studies (Bell et al., 2014; Suchodolski et al., 2015; Weese et al., 2015), relative abundance ranged from 50–80% with similar variability in Actinobacteria (0.11–9.9%), Bacteroidetes (0.15–33%), Fusobacteria (0–1.15%) and Proteobacteria (0–30%). Differences among studies also could reflect geographical variation, heterogeneity in diet, or variable exposure to other treatments known to impact the microbiome. Although cats in this study had not previously received antibiotics, probiotics or proton-pump inhibitors, data on potential exposure to them were not included in the other reports.
Because there was not a control group that received neither antibiotics nor probiotics, it is not possible to definitively ascribe long-term changes in the fecal microbiome and metabolome to antibiotic administration. Although the fecal microbiome of cats evolves with age, predominant bacterial groups and relative abundances appear relatively stable in colony cats based on evaluation of cats from one to nine months of age and eight to 14 years from the same colony (Jia et al., 2011a; Jia et al., 2011b). Further, we did not identify an association between the age of cats and the composition of the microbiome at baseline. As such, exposure to antibiotics is the most likely cause for the marked changes in fecal microbiome and metabolomic profiles over time. Interestingly, we also found no association between age and AAGS, in contrast to work in people (McFarland, 2008).
Conclusions
High-dose clindamycin therapy induces AAGS in 100% of cats, and severity of signs is often treatment-limiting. The incidence of vomiting was lower in cats concurrently administered a synbiotic, although the difference was not statistically significant. Antibiotic-induced dysbiosis and alterations in fecal metabolite profiles mirror those found in people. Changes in fecal metabolomic profiles included derangements in SCFA, bile acid, indole, sphingolipid, polyamine, benzoic acid, and cinnaminic acid metabolism. Changes in relative abundances of many OTUs and metabolite profiles persisted for >600 days after treatment discontinuation. Significant differences between synbiotic and placebo groups were noted for metabolites that affect immunomodulation, intestinal permeability and barrier function, colonization resistance, and oxidative stress. Further investigation is warranted to determine whether synbiotics blunt AAGS or the risk of antibiotic-associated metabolic derangements, such as obesity, in cats undergoing repeated antibiotic therapy.
Supplemental Information
Median (range) peak height of metabolites in feces collected at baseline (days 5–7), at the conclusion of antibiotic administration (days 26–28), and after a 603 day washout (days 631–633) from 16 healthy cats, 8 per group,+ that received 150 mg clindamycin with either a placebo or synbiotic PO once daily for 21 days. +Feces from one cat (synbiotic group) unavailable at days 631–633. * P-values adjusted based on the Benjamini and Hochberg False discovery rate (fdr).
Four cats were removed from the study for marked hyporexia (2 placebo, 1 synbiotic) or high baseline fecal scoring (1 synbiotic). FS, female spayed; MC, male castrated; FS, fecal score; 0, absent; 1, present
Funding Statement
This project was supported by the University of Tennessee, College of Veterinary Medicine Companion Animal Fund; University of Tennessee, College of Veterinary Medicine Center of Excellence in Livestock Diseases and Human Health Summer Research Program; and Nutramax Laboratories Veterinary Sciences, Inc., Lancaster, SC. The funders had no involvement in the design or performance of the study, writing of the manuscript, or the decision to submit the manuscript for publication. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
Two of the investigators (JCW, JSS) declare past receipt of honorariums from Nutramax Laboratories for public speaking and educational materials.
Author Contributions
Jacqueline C. Whittemore conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Jennifer E. Stokes conceived and designed the experiments, performed the experiments, analyzed the data, authored or reviewed drafts of the paper, approved the final draft.
Nicole L. Laia performed the experiments, authored or reviewed drafts of the paper, approved the final draft.
Joshua M. Price analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Jan S. Suchodolski performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Animal Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
This study was approved by the Institutional Animal Care and Use Committee of the University of Tennessee, Knoxville (protocol number 2169) and performed in compliance with “The Guide for the Care and Use of Laboratory Animals” in laboratory animal facilities that are AAALAC certified and exceed NIH standards of care.
Data Availability
The following information was supplied regarding data availability:
The microbiome dataset generated and analyzed during the current study is available in the SRA repository, accession number SRP08225 (https://www.ncbi.nlm.nih.gov/sra). The metabolomics data is available at the website of the NIH Common Fund’s Metabolomics Data Repository and Coordinating Center (supported by NIH grant, U01-DK097430), the Metabolomics Workbench (http://www.metabolomicsworkbench.org), where it has been assigned study number ST000979.
Clinical and metabolomic data have been supplied as Supplementary Files.
References
- Abecia et al. (2010).Abecia L, Hoyles L, Khoo C, Frantz N, McCartney AL. Effects of a novel galactooligosaccharide on the faecal microbiota of healthy and inflammatory bowel disease cats during a randomized, double-blind, cross-over feeding study. International Journal Of Probiotics and Prebiotics. 2010;5:61–68. [Google Scholar]
- Adams et al. (2005).Adams VJ, Campbell JR, Waldner CL, Dowling PM, Shmon CL. Evaluation of client compliance with short-term administration of antimicrobials to dogs. Journal of the American Veterinary Medical Association. 2005;226:567–574. doi: 10.2460/javma.2005.226.567. [DOI] [PubMed] [Google Scholar]
- Alam et al. (2016).Alam MA, Subhan N, Hossain H, Hossain M, Reza HM, Rahman MM, Ullah MO. Hydroxycinnamic acid derivatives: a potential class of natural compounds for the management of lipid metabolism and obesity. Nutrition & Metabolism. 2016;13:27. doi: 10.1186/s12986-016-0080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarellos & Landoni (2009).Albarellos GA, Landoni MF. Current concepts on the use of antimicrobials in cats. Veterinary Journal. 2009;180:304–316. doi: 10.1016/j.tvjl.2008.01.001. [DOI] [PubMed] [Google Scholar]
- AlShawaqfeh et al. (2017).AlShawaqfeh MK, Wajid B, Minamoto Y, Markel M, Lidbury JA, Steiner JM, Serpedin E, Suchodolski JS. A dysbiosis index to assess microbial changes in fecal samples of dogs with chronic inflammatory enteropathy. FEMS Microbiology Ecology. 2017;93:fix136–fix136. doi: 10.1093/femsec/fix136. [DOI] [PubMed] [Google Scholar]
- Aniwan et al. (2018).Aniwan S, Tremaine WJ, Raffals LE, Kane SV, Loftus JEV. Antibiotic use and new-onset inflammatory bowel disease in Olmsted County, Minnesota: a population-based case-control study. Journal of Crohn’s and Colitis. 2018;12:137–144. doi: 10.1093/ecco-jcc/jjx135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal et al. (2010).Bansal T, Alaniz RC, Wood TK, Jayaraman A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal et al. (2007).Bansal T, Englert D, Lee J, Hegde M, Wood TK, Jayaraman A. Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infection and Immunity. 2007;75:4597–4607. doi: 10.1128/IAI.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett (2002).Bartlett JG. Antibiotic-associated diarrhea. New England Journal of Medicine. 2002;346:334–339. doi: 10.1056/NEJMcp011603. [DOI] [PubMed] [Google Scholar]
- Bell et al. (2014).Bell ET, Suchodolski JS, Isaiah A, Fleeman LM, Cook AK, Steiner JM, Mansfield CS. Faecal microbiota of cats with insulin-treated diabetes mellitus. PLOS ONE. 2014;9:e108729. doi: 10.1371/journal.pone.0108729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagi et al. (2013).Biagi G, Cipollini I, Bonaldo A, Grandi M, Pompei A, Stefanelli C, Zaghini G. Effect of feeding a selected combination of galacto-oligosaccharides and a strain of Bifidobacterium pseudocatenulatum on the intestinal microbiota of cats. American Journal of Veterinary Research. 2013;74:90–95. doi: 10.2460/ajvr.74.1.90. [DOI] [PubMed] [Google Scholar]
- Chan et al. (2012).Chan YH, Fan MM, Fok CM, Lok ZL, Ni M, Sin CF, Wong KK, Wong SM, Yeung R, Yeung TT, Chow WC, Lam TH, Schooling CM. Antibiotics nonadherence and knowledge in a community with the world’s leading prevalence of antibiotics resistance: implications for public health intervention. American Journal of Infection Control. 2012;40:113–117. doi: 10.1016/j.ajic.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, Khan & Hannun (1992).Chao R, Khan W, Hannun YA. Retinoblastoma protein dephosphorylation induced by D-erythro-sphingosine. Journal of Biological Chemistry. 1992;267:23459–23462. [PubMed] [Google Scholar]
- Col, Fanale & Kronholm (1990).Col N, Fanale JE, Kronholm P. The role of medication noncompliance and adverse drug reactions in hospitalizations of the elderly. Archives of Internal Medicine. 1990;150:841–845. doi: 10.1001/archinte.1990.00390160093019. [DOI] [PubMed] [Google Scholar]
- De La Cochetière et al. (2010).De La Cochetière MF, Montassier E, Hardouin JB, Carton T, Le Vacon F, Durand T, Lalande V, Petit JC, Potel G, Beaugerie L. Human intestinal microbiota gene risk factors for antibiotic-associated diarrhea: perspectives for prevention. Risk factors for antibiotic-associated diarrhea. Microbial Ecology. 2010;59:830–837. doi: 10.1007/s00248-010-9637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai et al. (2009).Desai AR, Musil KM, Carr AP, Hill JE. Characterization and quantification of feline fecal microbiota using cpn60 sequence-based methods and investigation of animal-to-animal variation in microbial population structure. Veterinary Microbiology. 2009;137:120–128. doi: 10.1016/j.vetmic.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Dethlefsen et al. (2008).Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLOS Biology. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc et al. (2013).Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, Thomas G, Barbu V, Humbert L, Despras G, Bridonneau C, Dumetz F, Grill JP, Masliah J, Beaugerie L, Cosnes J, Chazouillères O, Poupon R, Wolf C, Mallet JM, Langella P, Trugnan G, Sokol H, Seksik P. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut. 2013;62:531–539. doi: 10.1136/gutjnl-2012-302578. [DOI] [PubMed] [Google Scholar]
- European Food Safety Authority (2012).European Food Safety Authority EFSA panel on additives and products or substances used in animal feed scientific. Opinion on the safety and efficacy of Prostora Max (Bifidobacterium animalis) as a feed additive for dogs. EFSA Journal. 2012;10:2964–2978. [Google Scholar]
- Fiehn et al. (2008).Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee DY, Lu Y, Moon S, Nikolau B. Quality control for plant metabolomics: reporting MSI-compliant studies. Plant Journal. 2008;53:691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- Fujiwara et al. (2013).Fujiwara N, Porcelli SA, Naka T, Yano I, Maeda S, Kuwata H, Akira S, Uematsu S, Takii T, Ogura H, Kobayashi K. Bacterial sphingophospholipids containing non-hydroxy fatty acid activate murine macrophages via Toll-like receptor 4 and stimulate bacterial clearance. Biochimica et Biophysica Acta. 2013;1831:1177–1184. doi: 10.1016/j.bbalip.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Garcia-Mazcorro et al. (2011).Garcia-Mazcorro JF, Lanerie DJ, Dowd SE, Paddock CG, Grutzner N, Steiner JM, Ivanek R, Suchodolski JS. Effect of a multi-species synbiotic formulation on fecal bacterial microbiota of healthy cats and dogs as evaluated by pyrosequencing. FEMS Microbiology Ecology. 2011;78:542–554. doi: 10.1111/j.1574-6941.2011.01185.x. [DOI] [PubMed] [Google Scholar]
- Goldenberg et al. (2015).Goldenberg JZ, Lytvyn L, Steurich J, Parkin P, Mahant S, Johnston BC. Probiotics for the prevention of pediatric antibiotic-associated diarrhea (Review) Cochrane Database of Systematic Reviews. 2015;2015(12):CD004827. doi: 10.1002/14651858.CD004827.pub4. [DOI] [PubMed] [Google Scholar]
- Greco (2011).Greco DS. Diagnosis and dietary management of gastrointestinal disease. 2011. https://www.purinaproplanvets.com/media/1202/gi_quick_reference_guide.pdf. [29 September 2017]. https://www.purinaproplanvets.com/media/1202/gi_quick_reference_guide.pdf
- Gustafsson et al. (1998).Gustafsson A, Lund-Tønnesen S, Berstad A, Midtvedt T, Norin E. Faecal short-chain fatty acids in patients with antibiotic-associated diarrhoea, before and after faecal enema treatment. Scandinavian Journal of Gastroenterology. 1998;33:721–727. doi: 10.1080/00365529850171666. [DOI] [PubMed] [Google Scholar]
- Hart et al. (2012).Hart ML, Suchodolski JS, Steiner JM, Webb CB. Open-label trial of a multi-strain synbiotic in cats with chronic diarrhea. Journal of Feline Medicine and Surgery. 2012;14:240–245. doi: 10.1177/1098612X11434386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel et al. (2012).Hempel S, Newberry SJ, Maher AR, Wang Z, Miles JN, Shanman R, Johnsen B, Shekelle PG. Probiotics for the prevention and treatment of antibiotic-associated diarrhea: a systematic review and meta-analysis. Journal of the American Medical Association. 2012;307:1959–1969. doi: 10.1001/jama.2012.3507. [DOI] [PubMed] [Google Scholar]
- Honneffer et al. (2015).Honneffer J, Guard B, Steiner JM, Suchodolski JS. Untargeted metabolomics reveals disruption within bile acid, cholesterol, and tryptophan metabolic pathways in dogs with idiopathic inflammatory bowel disease (abstr.) Gastroenterology. 2015;148(Suppl. 1):S–715. Abstract Mo1805. [Google Scholar]
- Honneffer et al. (2017).Honneffer JB, Steiner JM, Lidbury JA, Suchodolski JS. Variation of the microbiota and metabolome along the canine gastrointestinal tract. Metabolomics. 2017;13:26. doi: 10.1007/s11306-017-1165-3. [DOI] [Google Scholar]
- Hunter et al. (1995).Hunter RP, Lynch MJ, Ericson JF, Millas WJ, Fletcher AM, Ryan NI, Olson JA. Pharmacokinetics, oral bioavailability and tissue distribution of azithromycin in cats. Journal of Veterinary Pharmacology and Therapeutics. 1995;18:38–46. doi: 10.1111/j.1365-2885.1995.tb00549.x. [DOI] [PubMed] [Google Scholar]
- Jakobsson et al. (2010).Jakobsson HE, Jernberg C, Andersson AF, Sjolund-Karlsson M, Jansson JK, Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLOS ONE. 2010;5:e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferds et al. (2002).Jefferds MD, Laserson K, Fry AM, Roy S, Hayslett J, Grummer-Strawn L, Kettel-Khan L, Schuchat A, Team smotCfDCaPAA Adherence to antimicrobial inhalational anthrax prophylaxis among postal workers, Washington, D.C. 2001. Emerging Infectious Diseases. 2002;8:1138–1144. doi: 10.3201/eid0810.020331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia et al. (2011a).Jia J, Frantz N, Khoo C, Gibson GR, Rastall RA, McCartney AL. Investigation of the faecal microbiota of geriatric cats. Letters in Applied Microbiology. 2011a;53:288–293. doi: 10.1111/j.1472-765X.2011.03105.x. [DOI] [PubMed] [Google Scholar]
- Jia et al. (2011b).Jia J, Frantz N, Khoo C, Gibson GR, Rastall RA, McCartney AL. Investigation of the faecal microbiota of kittens: monitoring bacterial succession and effect of diet. FEMS Microbiology Ecology. 2011b;78:395–404. doi: 10.1111/j.1574-6941.2011.01172.x. [DOI] [PubMed] [Google Scholar]
- Johnston et al. (2011).Johnston BC, Goldenberg JZ, Vandvik PO, Sun X, Guyatt GH. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database of Systematic Reviews. 2011;2011(11):CD004827. doi: 10.1002/14651858.CD004827.pub3. [DOI] [PubMed] [Google Scholar]
- Kardas et al. (2005).Kardas P, Devine S, Golembesky A, Roberts C. A systematic review and meta-analysis of misuse of antibiotic therapies in the community. International Journal of Antimicrobial Agents. 2005;26:106–113. doi: 10.1016/j.ijantimicag.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Koeppel et al. (2006).Koeppel KN, Bertschinger H, Van Vuuren M, Picard J, Steiner JM, Williams D, Cardwell J. The use of a probiotic in captive cheetahs (Acinonyx jubatus) Journal of the South African Veterinary Association. 2006;77:127–130. doi: 10.4102/jsava.v77i3.359. [DOI] [PubMed] [Google Scholar]
- Kronman et al. (2012).Kronman MP, Zaoutis TE, Haynes K, Feng R, Coffin SE. Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics. 2012;130:e794–e803. doi: 10.1542/peds.2011-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau & Chamberlain (2016).Lau CSM, Chamberlain RS. Probiotics are effective at preventing Clostridium difficile-associated diarrhea: a systematic review and meta-analysis. International Journal of General Medicine. 2016;9:27–37. doi: 10.2147/IJGM.S98280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir-Wijnkoop et al. (2014).Lenoir-Wijnkoop I, Nuijten MJ, Craig J, Butler CC. Nutrition economic evaluation of a probiotic in the prevention of antibiotic-associated diarrhea. Frontiers in Pharmacology. 2014;5:13. doi: 10.3389/fphar.2014.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llor et al. (2013).Llor C, Hernandez S, Bayona C, Moragas A, Sierra N, Hernandez M, Miravitlles M. A study of adherence to antibiotic treatment in ambulatory respiratory infections. International Journal of Infectious Diseases. 2013;17:e168–172. doi: 10.1016/j.ijid.2012.09.012. [DOI] [PubMed] [Google Scholar]
- Löfmark et al. (2006).Löfmark S, Jernberg C, Jansson JK, Edlund C. Clindamycin-induced enrichment and long-term persistence of resistant Bacteroides spp. and resistance genes. Journal of Antimicrobial Chemotherapy. 2006;58:1160–1167. doi: 10.1093/jac/dkl420. [DOI] [PubMed] [Google Scholar]
- Manuja et al. (2013).Manuja R, Sachdeva S, Jain A, Chaudhary J. A comprehensive review on biological activities of P-hydroxy benzoic acid and its derivatives. International Journal of Pharmaceutical Sciences Review and Research. 2013;22:109–115. [Google Scholar]
- Marshall-Jones et al. (2006).Marshall-Jones ZV, Baillon ML, Croft JM, Butterwick RF. Effects of Lactobacillus acidophilus DSM13241 as a probiotic in healthy adult cats. American Journal of Veterinary Research. 2006;67:1005–1012. doi: 10.2460/ajvr.67.6.1005. [DOI] [PubMed] [Google Scholar]
- McFarland (2008).McFarland LV. Antibiotic-associated diarrhea: epidemiology, trends, and treatment. Future Microbiology. 2008;3:563–578. doi: 10.2217/17460913.3.5.563. [DOI] [PubMed] [Google Scholar]
- McFarland (2009).McFarland LV. Evidence-based review of probiotics for antibiotic-associated diarrhea and Clostridium difficile infections. Anaerobe. 2009;15:274–280. doi: 10.1016/j.anaerobe.2009.09.002. [DOI] [PubMed] [Google Scholar]
- McFarland (2016).McFarland LV. An observation on inappropriate probiotic subgroup classifications in the meta-analysis by Lau and Chamberlain. International Journal of General Medicine. 2016;9:333–336. doi: 10.2147/IJGM.S119970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen et al. (2016).Mikkelsen KH, Knop FK, Vilsbøll T, Frost M, Hallas J, Pottegård A. Use of antibiotics in childhood and risk of Type 1 diabetes: a population-based case-control study. Diabetic Medicine. 2016;34:272–277. doi: 10.1111/dme.13262. [DOI] [PubMed] [Google Scholar]
- Minamino et al. (2003).Minamino M, Sakaguchi I, Naka T, Ikeda N, Kato Y, Tomiyasu I, Yano I, Kobayashi K. Bacterial ceramides and sphingophospholipids induce apoptosis of human leukaemic cells. Microbiology. 2003;149:2071–2081. doi: 10.1099/mic.0.25922-0. [DOI] [PubMed] [Google Scholar]
- Minamoto et al. (2015).Minamoto Y, Otoni CC, Steelman SM, Büyükleblebici O, Steiner JM, Jergens AE, Suchodolski JS. Alteration of the fecal microbiota and serum metabolite profiles in dogs with idiopathic inflammatory bowel disease. Gut Microbes. 2015;6:33–47. doi: 10.1080/19490976.2014.997612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minois, Carmona-Gutierrez & Madeo (2011).Minois N, Carmona-Gutierrez D, Madeo F. Polyamines in aging and disease. Aging. 2011;3:716–732. doi: 10.18632/aging.100361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz et al. (2014).Muñoz EB, Dorado MF, Guerrero JE, Martinez FM. The effect of an educational intervention to improve patient antibiotic adherence during dispensing in a community pharmacy. Atencion Primaria. 2014;46:367–375. doi: 10.1016/j.aprim.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan et al. (2010).Narayan SS, Jalgaonkar S, Shahani S, Kulharni VN. Probiotics: current trends in the treatment of diarrhoea. Hong Kong Medical Journal. 2010;16(3):213–218. [PubMed] [Google Scholar]
- Ohland & MacNaughton (2010).Ohland CL, MacNaughton WK. Probiotic bacteria and intestinal epithelial barrier function. American Journal of Physiology: Gastrointestinal and Liver Physiology. 2010;298:G807–G819. doi: 10.1152/ajpcell.00094.2009. [DOI] [PubMed] [Google Scholar]
- Ouwehand et al. (2016).Ouwehand AC, Forssten S, Hibberd AA, Lyra A, Stahl B. Probiotic approach to prevent antibiotic resistance. Annals of Medicine. 2016;48:246–255. doi: 10.3109/07853890.2016.1161232. [DOI] [PubMed] [Google Scholar]
- Pagnini et al. (2010).Pagnini C, Saeed R, Bamias G, Arseneau KO, Pizarro TT, Cominelli F. Probiotics promote gut health through stimulation of epithelial innate immunity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:454–459. doi: 10.1073/pnas.0910307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechere et al. (2007).Pechere JC, Hughes D, Kardas P, Cornaglia G. Non-compliance with antibiotic therapy for acute community infections: a global survey. International Journal of Antimicrobial Agents. 2007;29:245–253. doi: 10.1016/j.ijantimicag.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Quigley (2012).Quigley EM. Prebiotics and probiotics: their role in the management of gastrointestinal disorders in adults. Nutrition in Clinical Practice. 2012;27:195–200. doi: 10.1177/0884533611423926. [DOI] [PubMed] [Google Scholar]
- Ramadan et al. (2014).Ramadan Z, Xu H, Laflamme D, Czarnecki-Maulden G, Li QJ, Labuda J, Bourqui B. Fecal microbiota of cats with naturally occurring chronic diarrhea assessed using 16S rRNA gene 454-pyrosequencing before and after dietary treatment. Journal of Veterinary Internal Medicine. 2014;28:59–65. doi: 10.1111/jvim.12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi et al. (2014).Rossi G, Pengo G, Caldin M, Piccionello AP, Steiner JM, Cohen ND, Jergens AE, Suchodolski JS. Comparison of microbiological, histological, and immunomodulatory parameters in response to treatment with either combination therapy with prednisone and metronidazole or probiotic VSL#3 strains in dogs with idiopathic inflammatory bowel disease. PLOS ONE. 2014;9:e94699. doi: 10.1371/journal.pone.0094699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert et al. (2014).Schubert AM, Rogers MAM, Ring C, Mogle J, Petrosino JP, Young VB, Aronoff DM, Schloss PD. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. mBio. 2014;5:e01021–01014. doi: 10.1128/mBio.01021-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott et al. (2016).Scott FI, Horton DB, Mamtani R, Haynes K, Goldberg DS, Lee DY, Lewis JD. Administration of antibiotics to children before age 2 years increases risk for childhood obesity. Gastroenterology. 2016;151:120–129. doi: 10.1053/j.gastro.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selinger et al. (2013).Selinger CP, Bell A, Cairns A, Lockett M, Sebastian S, Haslam N. Probiotic VSL#3 prevents antibiotic-associated diarrhoea in a double-blind, randomized, placebo-controlled clinical trial. Journal of Hospital Infection. 2013;84:159–165. doi: 10.1016/j.jhin.2013.02.019. [DOI] [PubMed] [Google Scholar]
- Šlapeta et al. (2015).Šlapeta J, Dowd SE, Alanazi AD, Westman ME, Brown GK. Differences in the faecal microbiome of non-diarrhoeic clinically healthy dogs and cats associated with Giardia duodenalis infection: impact of hookworms and coccidia. International Journal for Parasitology. 2015;45:585–594. doi: 10.1016/j.ijpara.2015.04.001. [DOI] [PubMed] [Google Scholar]
- Stokes, Price & Whittemore (2017).Stokes JE, Price JM, Whittemore JC. Randomized, controlled, crossover trial of prevention of clindamycin-induced gastrointestinal signs using a synbiotic in healthy research cats. Journal of Veterinary Internal Medicine. 2017;31:1406–1413. doi: 10.1111/jvim.14795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strompfová, Lauková & Ouwehand (2004).Strompfová V, Lauková A, Ouwehand AC. Selection of enterococci for potential canine probiotic additives. Veterinary Microbiology. 2004;100:107–114. doi: 10.1016/j.vetmic.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Suchodolski (2016a).Suchodolski JS. Diagnosis and interpretation of intestinal dysbiosis in dogs and cats. The Veterinary Journal. 2016a;S1090-0233:30033–30038. doi: 10.1016/j.tvjl.2016.04.011. [DOI] [PubMed] [Google Scholar]
- Suchodolski (2016b).Suchodolski JS. Diagnosis and interpretation of intestinal dysbiosis in dogs and cats. The Veterinary Journal. 2016b;215:30–37. doi: 10.1016/j.tvjl.2016.04.011. [DOI] [PubMed] [Google Scholar]
- Suchodolski et al. (2009).Suchodolski JS, Dowd SE, Westermarck E, Steiner JM, Wolcott RD, Spillman T, Harmoinen JA. The effect of the macrolide antibiotic tylosin on microbial diversity in the canine small intestine as demonstrated by massive parallel 16S rRNA gene sequencing. BMC Microbiology. 2009;9:210. doi: 10.1186/1471-2180-9-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchodolski et al. (2015).Suchodolski JS, Foster ML, Sohail MU, Leutenegger C, Queen EV, Steiner JM, Marks SL. The fecal microbiome in cats with diarrhea. PLOS ONE. 2015;10:e0127378. doi: 10.1371/journal.pone.0127378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchodolski et al. (2012).Suchodolski JS, Markel ME, Garcia-Mazcorro JF, Unterer S, Heilmann RM, Dowd SE, Kachroo P, Ivanov I, Minamoto Y, Dillman EM, Steiner JM, Cook AK, Toresson L. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLOS ONE. 2012;7:e51907. doi: 10.1371/journal.pone.0051907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajewska, Konarska & Kołodziej (2016).Szajewska H, Konarska Z, Kołodziej M. Probiotic bacterial and fungal strains: claims with evidence. Digestive Diseases. 2016;34:251–259. doi: 10.1159/000443359. [DOI] [PubMed] [Google Scholar]
- Van den Bemt et al. (2016).Van den Bemt PM, Chaaouit N, van Lieshout EM, Verhofstad MH. Noncompliance with guidelines on proton pump inhibitor prescription as gastroprotection in hospitalized surgical patients who are prescribed NSAIDs. European Journal of Gastroenterology and Hepatology. 2016;28:857–862. doi: 10.1097/MEG.0000000000000634. [DOI] [PubMed] [Google Scholar]
- Varughese, Vakil & Phillips (2013).Varughese CA, Vakil NH, Phillips KM. Antibiotic-associated diarrhea: a refresher on causes and possible prevention with probiotics—continuing education article. Journal of Pharmacy Practice. 2013;26:476–482. doi: 10.1177/0897190013499523. [DOI] [PubMed] [Google Scholar]
- Vesper et al. (1999).Vesper H, Schmelz EM, Nikolova-Karakashian MN, Dillehay DL, Lynch DV, Merrill AHJ. Sphingolipids in food and the emerging importance of sphingolipids to nutrition. Journal of Nutrition. 1999;129:1239–1250. doi: 10.1093/jn/129.7.1239. [DOI] [PubMed] [Google Scholar]
- Bureau of Veterinary Drugs (1995).Bureau of Veterinary Drugs Suspected drug adverse reactions reported to the Bureau of Veterinary Drugs. Canadian Veterinary Journal. 1995;36:246–249. [PMC free article] [PubMed] [Google Scholar]
- Videlock & Cremonini (2012).Videlock EJ, Cremonini F. Meta-analysis: probiotics in antibiotic-associated diarrhea. Alimentary Pharmacology and Therapeutics. 2012;35:1355–1369. doi: 10.1111/j.1365-2036.2012.05104.x. [DOI] [PubMed] [Google Scholar]
- Weese et al. (2015).Weese JS, Nichols J, Jalali M, Litster A. The rectal microbiota of cats infected with feline immunodeficiency virus infection and uninfected controls. Veterinary Microbiology. 2015;180:96–102. doi: 10.1016/j.vetmic.2015.08.012. [DOI] [PubMed] [Google Scholar]
- Woodmansey et al. (2004).Woodmansey EJ, McMurdo ME, Macfarlane GT, Macfarlane S. Comparison of compositions and metabolic activities of fecal microbiotas in young adults and in antibiotic-treated and non-antibiotic-treated elderly subjects. Applied and Environmental Microbiology. 2004;70:6113–6122. doi: 10.1128/AEM.70.10.6113-6122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia et al. (2015).Xia J, Sinelnikov I, Han B, Wishart DS. MetaboAnalyst 3.0—making metabolomics more meaningful. Nucleic Acids Research. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia & Wishart (2011).Xia J, Wishart DS. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nature Protocols. 2011;6:743–760. doi: 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Median (range) peak height of metabolites in feces collected at baseline (days 5–7), at the conclusion of antibiotic administration (days 26–28), and after a 603 day washout (days 631–633) from 16 healthy cats, 8 per group,+ that received 150 mg clindamycin with either a placebo or synbiotic PO once daily for 21 days. +Feces from one cat (synbiotic group) unavailable at days 631–633. * P-values adjusted based on the Benjamini and Hochberg False discovery rate (fdr).
Four cats were removed from the study for marked hyporexia (2 placebo, 1 synbiotic) or high baseline fecal scoring (1 synbiotic). FS, female spayed; MC, male castrated; FS, fecal score; 0, absent; 1, present
Data Availability Statement
The following information was supplied regarding data availability:
The microbiome dataset generated and analyzed during the current study is available in the SRA repository, accession number SRP08225 (https://www.ncbi.nlm.nih.gov/sra). The metabolomics data is available at the website of the NIH Common Fund’s Metabolomics Data Repository and Coordinating Center (supported by NIH grant, U01-DK097430), the Metabolomics Workbench (http://www.metabolomicsworkbench.org), where it has been assigned study number ST000979.
Clinical and metabolomic data have been supplied as Supplementary Files.