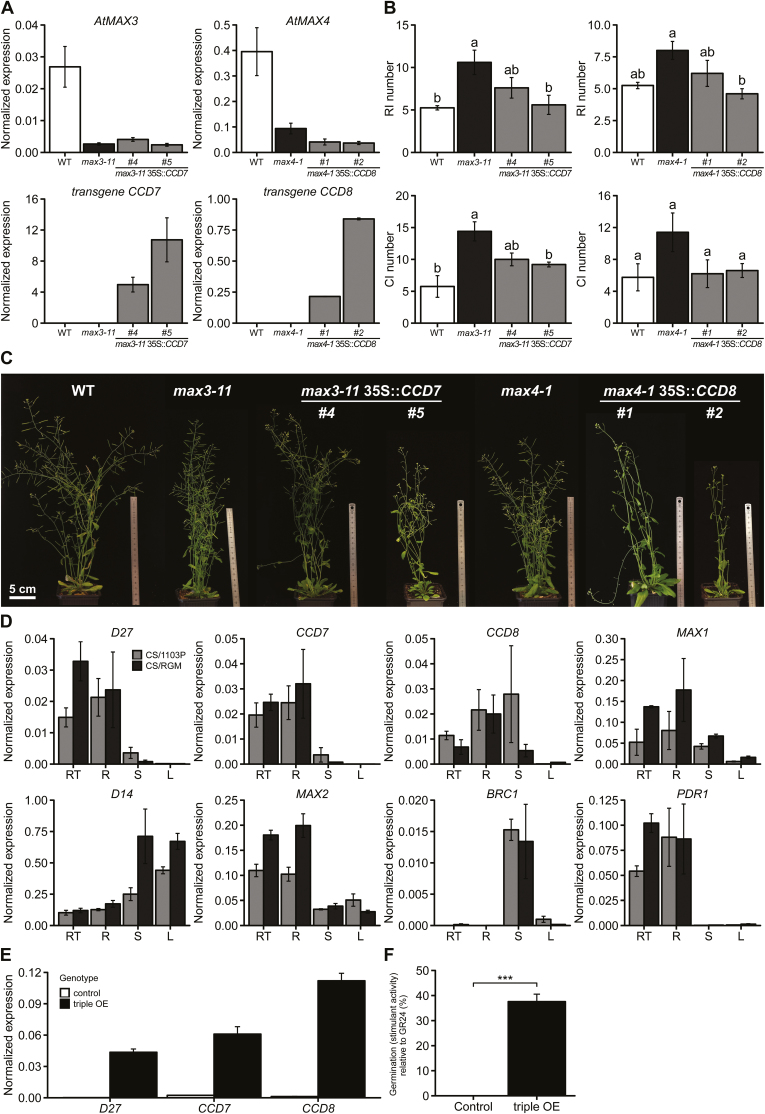
Fig. 3.
Functional characterization of grapevine SL-related genes. (A–C) Transformation of Arabidopsis max3-11 and max4-1 mutants with grapevine 35S::CCD7 or 35S::CCD8 genes, respectively. (A) Quantification of endogenous (AtMAX3 or AtMAX4) and transgene (CCD7 or CCD8) transcript levels in wild-type (Col-0), mutants, and two independent transgenic lines in the corresponding mutant background. RT–qPCR results from RNA isolated from shoots of 40-day-old seedlings are presented as normalized expression. Data are presented as means ±SE, n=3 individual plants. (B) Number of primary rosette branches (RI) and primary cauline branches (CI) of Col-0, max3-11, max4-1, and transgenic lines. Data are presented as means ±SE, n=10 individual plants. Letters above the bars indicate significant differences between genotypes as determined by Tukey’s multiple test (P<0.05). (C) Shoot branching phenotypes of Col-0, mutants, and transgenic lines. (D) Expression profiles of genes involved in SL biosynthesis (D27, CCD7, CCD8, MAX1), transport (PDR1), and signalling (D14, MAX2, BRC1) in different organs of 1-month-old CS/1103P and CS/RGM plants cultivated in a greenhouse with a low N content solution (0.8 mM NO3 –). L, leaves; R, roots; RT, root tips; S, stem. RT–qPCR results are presented as normalized expression. Data are presented as means ±SE, n=3 individual plants. (E, F) Overexpression of grapevine D27, CCD7, and CCD8 genes in 41B suspension cells. (E), Normalized expression of endogenous and transgenes in control and 35S::D27//35S::CCD7//35S::CCD8 (triple OE) transgenic cells. Data are presented as means ±SE, n=3 individual plants. (F) Activity of extracellular medium from control and transgenic (triple OE) cell suspensions (diluted 1:2) on P. ramosa seed germination. Germination-stimulant activities are represented as a percentage relative to the positive control GR24. Data are presented as means ±SE, n=9 (three biological replicates repeated three times). ***P<0.001, Student’s t-test.