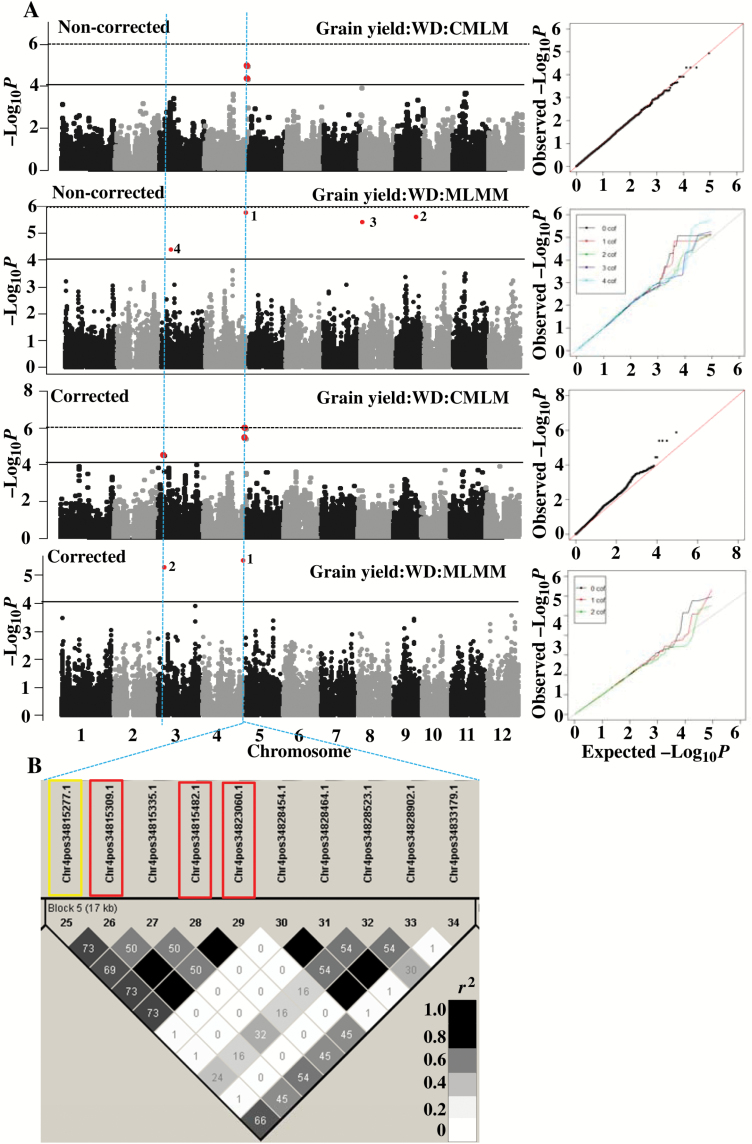
Fig. 4.
(A) GWAS results (Manhattan and quantile–quantile plot) detected through single-locus compressed mixed linear model (CMLM) and multi-locus mixed model (MLMM) for non-corrected and corrected (using days to flowering as covariate) grain yield in 2013 water-deficit stress (WD) conditions. Significant SNPs in the Manhattan plot of MLMM are numbered according to the order in which they were included as a cofactor in the regression model. (B) Identified LD block (17 kb) based on r2 value between SNPs on chromosome 4 and the colour intensity of the box on the LD plot corresponds with r2 (multiplied by 100) according to legend. Significant SNP marked by first rectangle was detected by CMLM and MLMM and the next three rectangles only by CMLM approach.