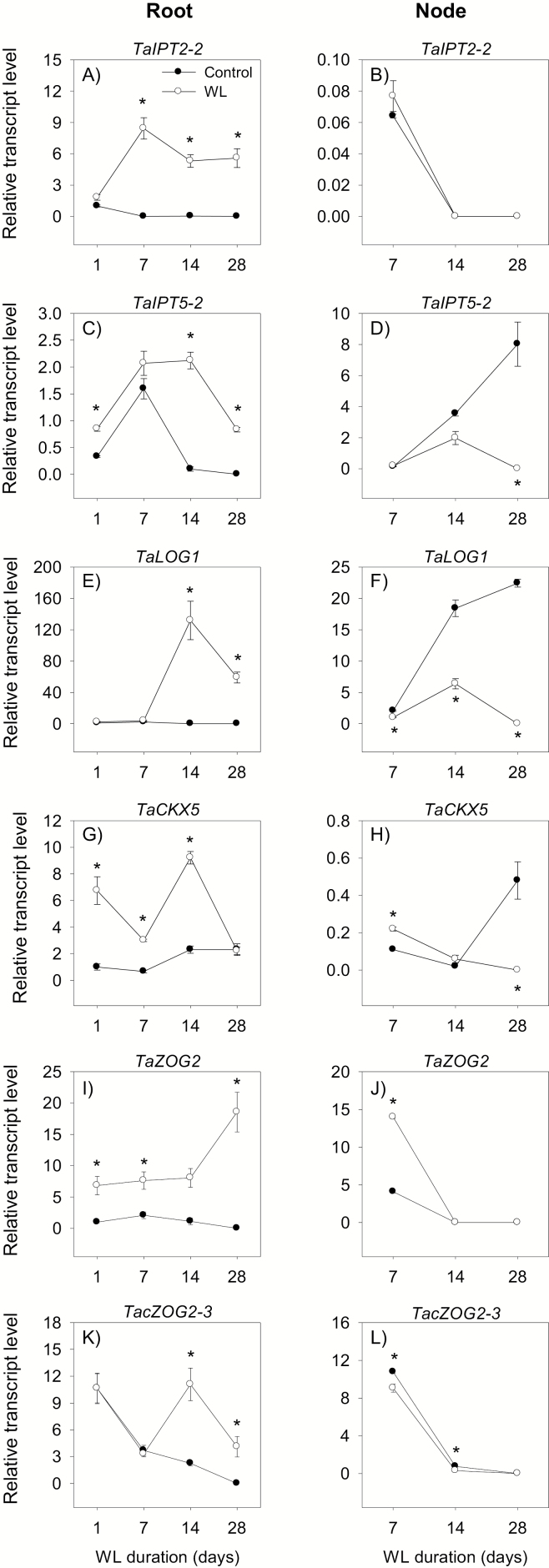
Fig. 7.
Expression of cytokinin metabolism genes. Relative transcript levels of IPT2-2 (A and B), IPT5-2 (C and D), LOG1 (E and F), CKX5 (G and H), ZOG2 (I and J), and cZOG2-3 (K and L) in the root tissue waterlogged for 1, 7, 14, and 28 d (A, C, E, G, I, and K) and stem node tissue waterlogged for 7, 14, and 28 d (B, D, F, H, J, and L), and their respective controls. Transcript levels of IPT genes, LOG1, CKX5, and ZOG genes in both tissues were determined as described in Fig. 3, and are expressed relative to the transcript levels of IPT2-2, LOG1, CKX5, and ZOG2 in control roots at 1 d after the start of waterlogging, respectively, which were arbitrarily set a value of 1. Other data descriptions are as indicated in Fig. 3. Expression levels of LOG3 in both tissues, and IPT2-2 and LOG5 in the stem node were either minimal or undetectable irrespective of growth conditions, as shown in Supplementary Table S3.