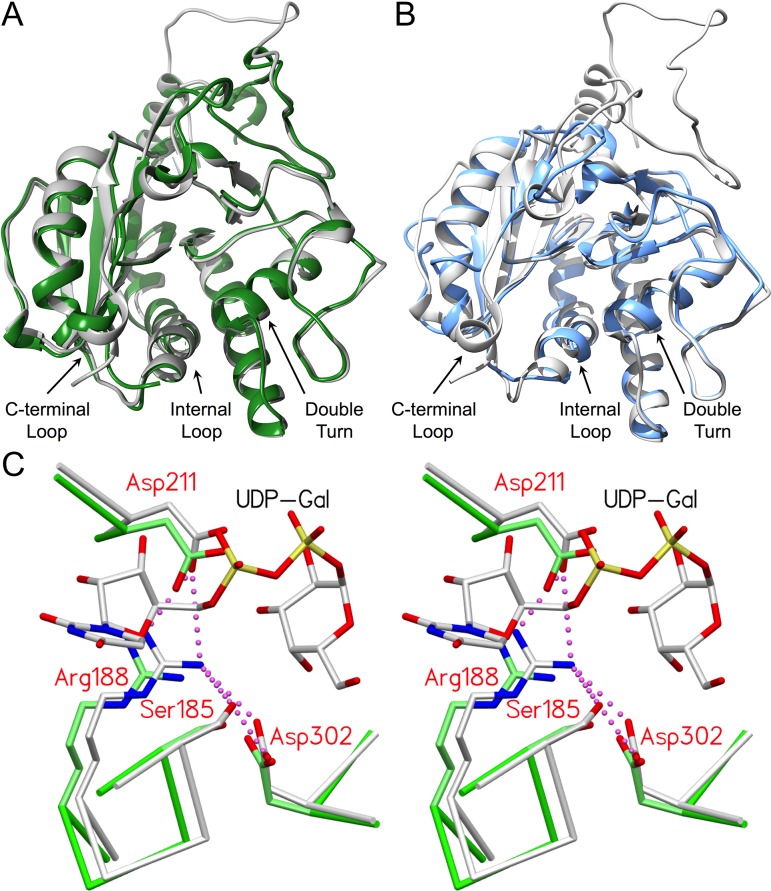
Fig. 1.
GTA and GTB are structurally homologous with family 6 GTs α3GalT and BoGT6a and share a number of conserved, equivalent residues critical to forming and stabilizing the salt bridge network. (A) Overlap of α3GalT (green, PDB code 1G93) with the catalytically competent closed state of the chimeric enzyme AABB (gray; PDB code 2RJ7; 0.77 Å RMSD). (B) Overlap of BoGT6a (blue; PDB code 4AYJ; 1.08 Å RMSD) with AABB (gray; PDB code 2RJ7). (C) Stereoview of the superposition of closed chimeric enzyme AABB (gray; PDB code 2RJ7) and α3GT (green; PDB code 1G93) showing the equivalent positions of active site residues Ser185/199, Arg188/202, Asp211/225 and Asp302/316 (GTB side chains labeled) with UDP-Gal bound in the catalytically competent “tucked under” conformation #1. Salt bridge interactions are depicted as magenta dashed spheres. Distances for all interactions are in Tables IV and V.