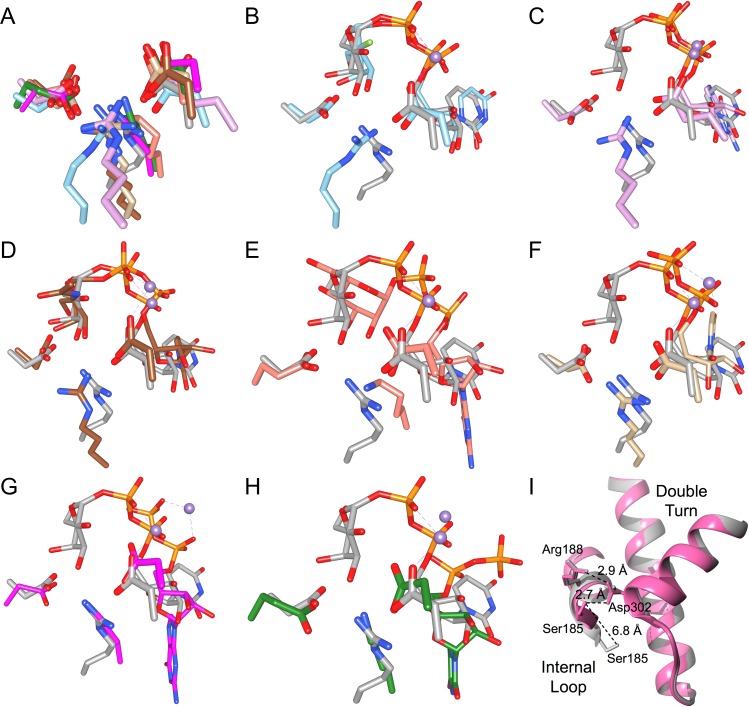
Fig. 4.
The salt bridge interactions formed in the closed enzyme state represent a structurally conserved feature found in representative structures from each retaining GT-A fold family. (A) Overlap of salt bridge residues Arg188, Asp211 and Asp302 from chimeric AABB in gray (PDB code 2RJ7) with corresponding residues (listed in Table VI) in representative structures from each retaining GT-A fold family using the following color scheme: GT-8 in sky blue (PDB code 1GA8), GT-15 in plum (PDB code 1SP4), GT-27 in brown (PDB code 4D0T), GT-55 in salmon (PDB code 2ZU8), GT-64 in tan (PDB code 1ON8), GT-78 in deep pink (PDB code 2Y4L) and GT-81 in forest green (PDB code 4DEC). In (B-H) AABB again is shown in gray (PDB code 2RJ7) and is separately overlapped with each GT-A fold family representative, which collectively are colored with cyan for carbon, phosphorous and manganese. Overlaps are between AABB and a representative structure (listed in Table VI) of (B) GT-8, (C) GT-15, (D) GT-27, (E) GT-55, (F) GT-64, (G) GT-78 and (H) GT-81. Atoms are colored by element with oxygen red, nitrogen blue, fluorine light green, carbon as indicated above, phosphorous orange except where otherwise indicated and manganese medium-purple except where otherwise indicated. Color naming is consistent with the program USCF Chimera. (I) Overlap of AABB (pink, PDB code 2RJ7) with the semi-open GTB (gray, PDB code 5C1L) showing the >6 Å shift of internal loop residue Ser185 that occurs upon substrate binding and salt bridge reorganization.