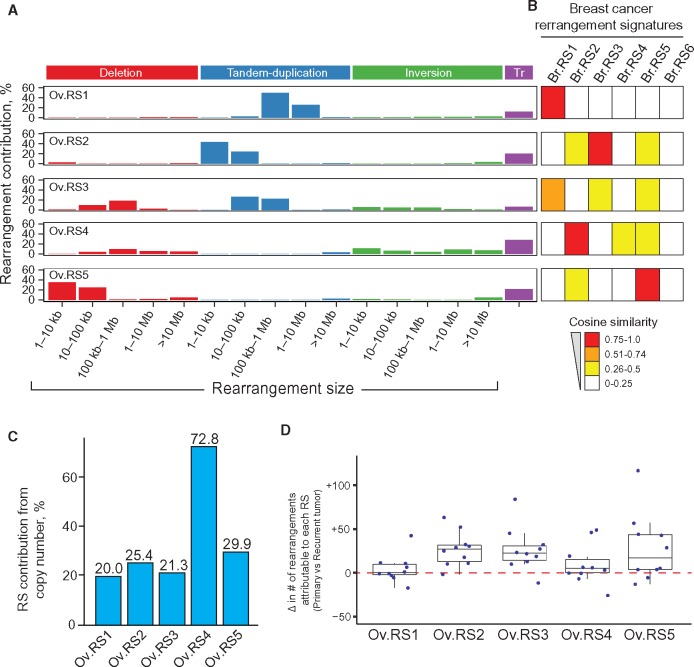
Figure 2.
Five genomic rearrangement signatures identified from high-grade serous ovarian cancer (HGSOC) tumor whole-genome sequencing data. A) The five rearrangement signatures include distinct contributions from rearrangement classes and are dissimilar from one another (mean pairwise cosine similarity = 0.28). No signature had a substantial contribution from clustered rearrangements (mean signature contribution from clustered rearrangements < 0.01%), and thus for clarity clustered rearrangement contribution is not shown. B) The HGSOC rearrangement signatures were compared with previously identified breast cancer rearrangement signatures using cosine similarity, as represented by heatmap values (8). Ov.RS1, Ov.RS2, Ov.RS4, and Ov.RS5 were similar to breast cancer rearrangement signatures (cosine similarity > 0.90), but in contrast Ov.RS3 did not have high similarity with any breast cancer rearrangement signature (maximum cosine similarity = 0.70). C) The percent contribution from copy number–neutral events for each of the five HGSOC rearrangement signatures. D) Change in the number of rearrangements attributable to each rearrangement signature, as compared with 10 matched primary and recurrent tumor pairs from the same patients. RS = rearrangement signature; Tr = inter-chromosomal translocation.