Abstract
Fructosamine and glycated albumin are potentially useful alternatives to hemoglobin A1c (HbA1c) as diabetes biomarkers. The genetic determinants of fructosamine and glycated albumin, however, are unknown. We performed genome-wide association studies of fructosamine and glycated albumin among 2,104 black and 7,647 white participants without diabetes in the Atherosclerosis Risk in Communities (ARIC) Study and replicated findings in the Coronary Artery Risk Development in Young Adults (CARDIA) study. Among whites, rs34459162, a novel missense single nucleotide polymorphism (SNP) in RCN3, was associated with fructosamine (P = 5.3 × 10−9) and rs1260236, a known diabetes-related missense mutation in GCKR, was associated with percent glycated albumin (P = 5.9 × 10−9) and replicated in CARDIA. We also found two novel associations among blacks: an intergenic SNP, rs2438321, associated with fructosamine (P = 6.2 × 10−9), and an intronic variant in PRKCA, rs59443763, associated with percent glycated albumin (P = 4.1 × 10−9), but these results did not replicate. Few established fasting glucose or HbA1c SNPs were also associated with fructosamine or glycated albumin. Overall, we found genetic variants associated with the glycemic information captured by fructosamine and glycated albumin as well as with their nonglycemic component. This highlights the importance of examining the genetics of hyperglycemia biomarkers to understand the information they capture, including potential glucose-independent factors.
Introduction
Diabetes is defined by elevated blood glucose levels (hyperglycemia). Hemoglobin A1c (HbA1c) is formed as glucose binds to hemoglobin molecules within erythrocytes and is the standard clinical measure of chronic hyperglycemia used to diagnose and monitor diabetes (1). However, factors related to the nonglycemic portion of HbA1c such as erythrocyte turnover and hemoglobin characteristics can affect HbA1c values (2).
There is growing interest in fructosamine and glycated albumin, additional biomarkers of hyperglycemia that demonstrate associations with diabetes risk and complications similar to HbA1c (3–14). Fructosamine measures total serum protein bound to glucose. Glycated albumin is expressed as a percentage of serum albumin, the most abundant serum protein. Both biomarkers reflect glucose exposure over a shorter period of time (2–4 weeks) than HbA1c (2–3 months) (15). Fructosamine may be used to monitor glycemic control in clinical situations where HbA1c is problematic, such as in the setting of anemia or hemoglobinopathies (2). While glycated albumin is not frequently used in the U.S., it is widely used in Japan and other countries as a complement to HbA1c to monitor short-term glycemic control (16).
The genetics of HbA1c and glucose have been well studied; however, genetic factors that influence fructosamine and glycated albumin are uncharacterized. Of the known genome-wide association study (GWAS) variants associated with fasting glucose and HbA1c in European ancestry cohorts, few loci are associated with both (17). Many HbA1c variants are in genes related to hematological factors rather than glucose metabolism (18–20), while fasting glucose variants are in genes involved in glucose metabolism (although these variants are not all associated with diabetes) (21–23). This lack of overlap suggests that some underlying genetic variants are specific to particular biomarkers of hyperglycemia rather than to type 2 diabetes. Understanding the genetic determinants of fructosamine and glycated albumin should help in the interpretation of these tests and possibly extend our understanding of the pathophysiology of glucose metabolism. In particular, comparing the genetic overlap between different measures of glycemia may provide insight into the contributions of glycemic versus nonglycemic gene variants, i.e., to what extent genetic factors operate via pathways directly relevant to diabetes pathophysiology (“glycemic”) or operate via glycemic-independent pathways that do not influence glucose metabolism or diabetes risk (“nonglycemic,” such as the hematological variants associated with HbA1c). If nonglycemic genetic variants strongly impact fructosamine and glycated albumin, this may need to be taken into account in the interpretation of these biomarkers as measures of hyperglycemia.
We conducted GWAS of fructosamine and glycated albumin in blacks and whites in the Atherosclerosis Risk in Communities (ARIC) Study. We also compared previously identified genetic determinants for HbA1c and fasting glucose with fructosamine and glycated albumin to identify common genetic factors related to glucose metabolism and those that may be distinct to fructosamine and glycated albumin.
Research Design and Methods
Study Population
The ARIC Study is an ongoing prospective cohort of 15,792 participants initiated in 1987 (24). Participants were middle-aged adults recruited from four U.S. communities (Jackson, Mississippi; Forsyth County, North Carolina; Washington County, Maryland; and suburban Minneapolis, Minnesota). All study participants provided written informed consent, and study protocols were approved by the relevant institutional review boards.
In the current study, we included 9,751 participants (7,647 whites and 2,104 blacks) who attended visit 2 (1990–1992), consented for use of DNA, did not have diagnosed diabetes (self-reported diagnosis or use of diabetes medications), had valid data on fructosamine and glycated albumin, and had genotyping data meeting quality control criteria (Supplementary Fig. 1). Individuals with diagnosed diabetes were excluded to avoid potential bias caused by altered glucose levels as a result of diabetes treatment.
Genotyping
ARIC Study participants were genotyped using the Affymetrix 6.0 array and imputed separately by race using IMPUTE2 (25) with the 1000 Genomes Project phase 1 (March 2012) reference panel. Quality control excluded individuals based on single nucleotide polymorphism (SNP) mismatch, high discordance with previous TaqMan assay genotypes, genetic outlier status, and relatedness. SNPs with IMPUTE info score <0.8 or minor allele frequency (MAF) <0.05 were excluded. Only autosomal variants (on chromosomes 1–22) were considered. Principal components analysis was used to estimate population substructure with EIGENSTRAT (26).
Glycemic Markers
Fructosamine (Roche Diagnostics, Indianapolis, IN) and glycated albumin and serum albumin (GA-L; Asahi Kasei Pharma Corporation, Tokyo, Japan) were measured in 2012–2013 using a Roche Modular P800 system from serum collected at visit 2 and stored at −70°C (3). Percent glycated albumin was calculated per the manufacturer’s protocol: [(glycated albumin concentration in g/dL/serum albumin concentration in g/dL)*100/1.14] + 2.9. We also examined total glycated albumin (g/dL) as well as serum albumin (g/dL) to help distinguish genetic factors specific to serum protein concentration versus hyperglycemia.
Serum glucose was measured on the Roche Hitachi 911 analyzer using the hexokinase method (Roche Diagnostics), and HbA1c was measured from whole blood stored at −70°C using high-performance liquid chromatography, standardized to the assay used in the Diabetes Control and Complications Trial (DCCT) (27).
Statistical Analysis
GWAS in blacks and whites were conducted using SNPTEST v2 (28) for all glycemic biomarkers using imputed allele dosage and controlling for age, sex, field center, and the first 10 principal components under an additive genetic model. Fructosamine and glycated albumin (both percent (%) and total [g/dL]) were transformed on the natural log scale; therefore, the effect sizes are the change in the natural log of the biomarker per each additional risk allele. Exponentiating the effect sizes thus corresponds to the percent higher or lower biomarker levels per additional risk allele. Fasting glucose and HbA1c were not transformed. To identify additional independent SNPs associated with the traits, we performed conditional analyses for genome-wide significant findings. Using fructosamine, percent glycated albumin, and total glycated albumin as the dependent variable and the index SNP (SNP with the lowest P value in a region showing a genome-wide significant association) as a covariate, we evaluated the association between other SNPs with a MAF ≥1% within 250 kB of the index SNP or between recombination hotspots surrounding the index SNP. To estimate percentage of variance explained by each SNP, we used the equation Ri2 = bi2 × var(SNPi)/var(y) where bi = the effect size of the association between the SNPi and the phenotype y, var(SNPi) is 2 × MAFSNPi × (1-MAFSNPi) and var(y) is the variance in the phenotype (29,30). We meta-analyzed across ancestries using a random-effects model by GWAMA (31).
In sensitivity analyses, we performed GWAS excluding case subjects with undiagnosed diabetes (participants with fasting glucose ≥126 mg/dL or nonfasting glucose >200 mg/dL). To further evaluate the genetic variants pertaining to serum protein levels rather than hyperglycemia, we evaluated the top fructosamine and glycated albumin SNPs for association with total glycated albumin and with serum albumin (transformed on the natural log scale). To determine the extent to which glycemic biomarkers shared genetics, we used the program LDSC, which calculated genetic correlations between the biomarkers from ARIC summary statistics by taking advantage of the LD structure (variants in LD with more SNPs are more likely to have larger effect sizes, which extends to the product of correlated traits). We used precomputed LD scores from 1000 Genomes European data (32). Genetic correlations are not bounded to [–1,1], and estimates outside of this range indicate strong genetic correlations.
Replication
Significant associations between genetic variants and fructosamine and percent glycated albumin in ARIC were evaluated for replication in the Coronary Artery Risk Development in Young Adults (CARDIA) cohort, a prospective cohort study initiated in 1985 to evaluate risk factors for heart disease among unrelated young adults (33).
Serum specimens from 2005 to 2006 were stored at –70°C and used to analyze glycated albumin and fructosamine in 2014 using a Roche COBAS 6000 chemistry analyzer (Roche Diagnostics). Glycated albumin and fructosamine were measured using the same assays used in ARIC (glycated albumin by Lucica GA-L from Asahi Kasei Pharma Corporation and fructosamine via Roche Diagnostics).
Genotyping was performed using the Affymetrix 6.0 array. Standard quality control metrics were applied, and imputation to HapMap Phase II, Build 36, Release 22 was done using MACH (34). Genetic, covariate, fructosamine, and glycated albumin data were available on 1,304 whites and 608 blacks. Individuals with diabetes (current use of glucose-lowering medications or fasting glucose ≥126 mg/dL) were excluded from analysis. Linear regression analyses stratified by race were done for the association between significant ARIC SNPs and natural log–transformed fructosamine and percent glycated albumin adjusted for age, sex, field center, and the first three principal components.
Statistical analysis was done using SAS v9.4 (SAS Institute, Cary, NC) (data manipulation) and ProbABELv0.2 (35). If the ARIC SNP was not available in the CARDIA data set, we determined whether proxy SNPs in linkage disequilibrium (LD) with the ARIC SNP (r2 > 0.7 from 1000 Genomes phase 3v5 European population, GRCh37 assembly using Single Nucleotide Polymorphisms Annotator (SNiPA) [36]) were available and, if so, analyzed the associations with the proxy SNP. We considered a Bonferroni-corrected one-sided P value threshold of <0.05 (0.1/2 SNPs per race) for replication significance. We meta-analyzed ARIC and CARDIA results using fixed-effects inverse variance weighted model by METAL (37).
Candidate SNP Analysis
We additionally evaluated previously identified fasting glucose (n = 41) and HbA1c (n = 46) candidate SNPs from the National Human Genome Research Institute (NHGRI)-EBI Catalog of published GWAS using the search terms “fasting glucose” and “HbA1c” (http://www.ebi.ac.uk/gwas/ as of 14 December 2017) for association with fructosamine and percent glycated albumin in ARIC. SNPs were included if they were discovered in European ancestry cohorts, were genome-wide significant (P < 5 × 10−8), and were not in LD with each other (r2 < 0.2, using SNiPA) (36). For the candidate SNP analyses, we used a study-wide significance threshold: two traits (fructosamine and glycated albumin), two races (black and white), and the number of candidate SNPs for each trait: P < 4.6 × 10−4 (0.05/(2*2*27)) for fasting glucose and P < 2.6 × 10−4 (0.05/(2*2*49)) for HbA1c.
We additionally performed analyses controlling for known fasting glucose and HbA1c SNPs with P < 0.05 in ARIC, compiled into scores calculated as the sum of the number of risk alleles, weighted by the effect size in ARIC among whites.
Comparison of Variance Explained
To compare the influence of glycemic and nonglycemic genetic variants on fructosamine and glycated albumin to that of HbA1c, we calculated the percent phenotypic variance explained by the SNPs in our study to those from published results of HbA1c. In addition, we calculated variance explained by known fasting glucose and albumin SNPs (identified by the same criteria used for fasting glucose and HbA1c). Percent variance explained was calculated using the equation described above.
Results
Overall, 7,647 whites and 2,104 blacks from ARIC and 1,304 whites and 608 blacks from CARDIA were included in this study (Supplementary Table 1). Mean age in ARIC (56–57 years) was higher than in CARDIA (45–46 years). The cohorts had similar distribution of sex. CARDIA had a greater percentage of black participants and lower mean values for each measure of glycemia as compared with ARIC.
We identified four genome-wide significant loci in ARIC, two associated with fructosamine and two with percent glycated albumin. Three of these variants, rs34459162 intronic to RCN3, rs2438321 (intergenic), and rs59443763 intronic to PRKCA have not previously been reported to be associated with any glycemic traits in humans (Table 1). None of the analyses showed evidence for inflation (Supplementary Figs. 2–7).
Table 1.
Genome-wide significant loci for fructosamine and percent glycated albumin
| SNP | Chr | Closest gene | A1/A2† | Outcome | Race | ARIC* |
CARDIA* |
ARIC+CARDIA* |
ARIC White+Black* |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 freq | β (SE)‡ | % diff§ | P value | % variance explained | A1 freq | β (SE)‡ | % diff§ | P value | β (SE)‡ | % diff§ | P value | β (SE)‡ | % diff§ | P value | ||||||
| rs34459162|| |
19 |
RCN3 |
C/T |
Fructosamine (μmol/L) |
White |
0.08 |
−0.02 (0.003) |
−2% |
5.3E-9 |
0.6% |
0.09 |
−0.02 (0.014) |
−2% |
0.09 |
−0.02 (0.003) |
−2% |
4.9E-9 |
— |
— |
— |
| rs1260326 |
2 |
GCKR |
T/C |
Percent glycated albumin (%) |
White |
0.41 |
−0.01 (0.002) |
−1% |
5.9E-9 |
0.3% |
0.43 |
−0.01 (0.004) |
−1% |
0.04 |
−0.01 (0.002) |
−1% |
2.3E-8 |
−0.007 (0.005) |
−0.7% |
0.14 |
| rs2438321|| |
11 |
CNTN5 |
G/A |
Fructosamine (μmol/L) |
Black |
0.11 |
0.03 (0.006) |
3% |
6.2E-9 |
1.8% |
0.06 |
0.006 (0.011) |
0.6% |
0.57 |
0.03 (0.005) |
3% |
2.9E-8 |
0.02 (0.02) |
2% |
0.37 |
| rs59443763 | 17 | PRKCA | C/T | Percent glycated albumin (%) | Black | 0.06 | 0.05 (0.009) | 5% | 4.1E-9 | 2.0% | — | — | — | — | — | — | — | — | — | — |
Chr, chromosome; diff, difference; freq, frequency.
*ARIC: N = 7,647 whites, 2,104 blacks; CARDIA: N = 1,304 whites, 608 blacks; ARIC+CARDIA is a meta-analysis across the cohorts; ARIC White+Black is a meta-analysis across the ancestries in ARIC.
†A1 is the minor allele in whites.
‡Mean change in ln(outcome) for each additional A1 allele.
§Percent higher or lower levels of the outcome for each additional copy of the minor allele, calculated as eβ *100.
||rs34459162 and rs2438321 not available in CARDIA data set; evaluated proxy SNPs rs8105626 and rs35256014, respectively, in perfect LD (r2 = 1 with ARIC SNP).
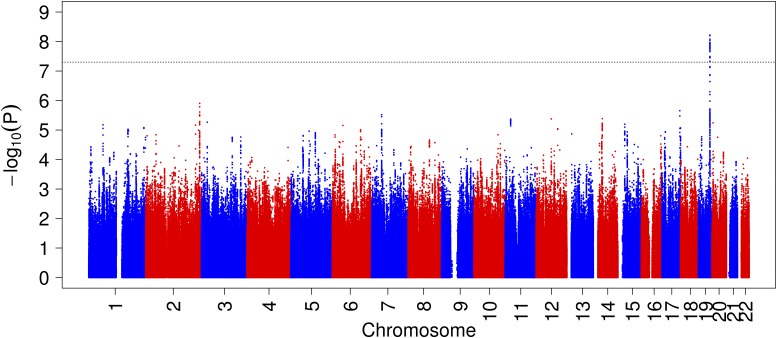
Among whites, rs34459162 (MAF = 0.08), a missense SNP in RCN3 on chromosome 19, was significantly associated with 1.8% lower fructosamine per minor allele (P = 5.3 × 10−9, variance explained = 0.6%) (Table 1 and Figs. 1 and 2). This SNP was also associated with total glycated albumin (P = 3.8 × 10−8) (Table 2). The association with percent glycated albumin approached genome-wide significance (P = 7.3 × 10−8), but this SNP was not associated with fasting glucose or HbA1c (Table 2). A proxy for rs34459162 (rs8105626, in r2 = 1 rs34459162) was not associated in CARDIA for association with fructosamine (P = 0.09), although the effect sizes were identical and meta-analysis across the cohorts was significant (P = 4.9 × 10−9) (Table 1). Conditional analysis in ARIC showed that the additional 63 significant SNPs in the region became nonsignificant after conditioning on rs34459162. In blacks, rs34459162 did not meet the info score >0.8 threshold and thus was not analyzed (Table 2).
Figure 1.
Manhattan plot for GWAS of fructosamine (log transformed) in whites (N = 7,647).
Figure 2.
Regional association plot for rs34459162 and fructosamine (log transformed) among whites (included SNPs with MAF ≥5% and imputation quality ≥0.8, insertions and deletions excluded).
Table 2.
Genome-wide significant loci for fructosamine and percent glycated albumin and their association with total glycated albumin, fasting glucose, and HbA1c in ARIC
| SNP |
Closest gene |
A1/A2‡ |
Race |
1000 Genomes A1 freq§ |
A1 freq |
Fructosamine (μmol/L) |
Percent glycated albumin (%) |
Total glycated albumin (g/dL) |
Fasting glucose (mg/dL) |
HbA1c (%) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β (SE) | P value | β (SE) | P value | β (SE) | P value | β (SE) | P value | β (SE) | P value | ||||||
| rs34459162 |
RCN3 |
C/T |
White |
0.08 |
0.08 |
-0.02 (0.003) |
5.3E-9 |
−0.02 (0.004) |
7.3E-8 |
−0.03 (0.005) |
3.8E-8 |
0.26 (0.56) |
0.64 |
−0.02 (0.02) |
0.25 |
| Black |
— |
— |
— |
— |
— |
— |
— |
— |
— |
— |
— |
— |
|||
| rs1260326 |
GCKR |
T/C |
White |
0.43 |
0.41 |
−0.003 (0.002) |
0.02 |
−0.01 (0.002) |
5.9E-9 |
−0.01 (0.003) |
1.3E-5 |
−1.13 (0.28) |
4.5E-5 |
−0.01 (0.008) |
0.09 |
| Black |
0.10 |
0.14 |
0.0005 (0.005) |
0.93 |
−0.0003 (0.006) |
0.96 |
0.001 (0.009) |
0.9 |
−0.53 (1.16) |
0.64 |
−0.01 (0.04) |
0.76 |
|||
| rs2438321 |
CNTN5 |
G/A |
White |
0.30 |
0.24 |
−0.001 (0.002) |
0.44 |
0.0005 (0.002) |
0.82 |
−0.001 (0.003) |
0.71 |
−0.21 (0.32) |
0.52 |
0.001 (0.01) |
0.95 |
| Black |
0.11 |
0.11 |
0.03 (0.006) |
6.2E-9 |
0.03 (0.007) |
6.4E-5 |
0.05 (0.009) |
2.0E-6 |
4.19 (1.24) |
8.3E-4 |
0.14 (0.04) |
6.8E-4 |
|||
| rs59443763 | PRKCA | C/T | White |
0.005 |
— |
— |
— |
— |
— |
— |
— |
— |
— |
— |
— |
| Black | 0.09 | 0.06 | 0.04 (0.008) | 9.4E-7 | 0.05 (0.009) | 4.1E-9 | 0.06 (0.01) | 5.8E-7 | 7.2 (1.65) | 9.6E-6 | 0.18 (0.05) | 8.3E-4 | |||
freq, frequency. Genome-wide significant results are in boldface type. Fructosamine, glycated albumin percent, and total glycated albumin are log transformed.
‡A1 is the minor allele in whites.
§1000 Genomes populations: white, Utah residents (CEPH) with Northern and Western European ancestry (CEU); black, Americans of African ancestry in Southwest U.S. (ASW).
Among whites, rs1260326 (also known as rs343480), a known missense mutation in GCKR on chromosome 2 (MAF = 0.41), was significantly associated with 1.1% lower levels of percent glycated albumin per minor allele (P = 5.3 × 10−9, variance explained = 0.3%) (Supplementary Figs. 8 and 9) (Table 1). The association with percent glycated albumin was also significant in CARDIA (P = 0.04), with similar percent difference (0.8% lower per minor allele) and genome-wide significant meta-analysis results (2.3 × 10−8) (Table 1). The conditional analysis did not reveal additional independent signals in this region. This SNP was not associated with any biomarker among blacks (MAF = 0.14) (Table 2 and Supplementary Table 2), but power was limited, and the meta-analysis across ancestries was not significant (Table 1 and Supplementary Fig. 10).
Among blacks, rs2438321 on chromosome 11 (MAF = 0.11) was associated with 3.5% higher levels of fructosamine per minor allele at a genome-wide significant level (P = 6.2 × 10−9, variance explained = 1.8%) (Table 1 and Supplementary Figs. 11 and 12) and approached significance with percent glycated albumin (P = 6.4 × 10−5) and total glycated albumin (P = 2.0 × 10−6) (Table 2). rs2438321 was not associated with HbA1c in blacks and was not associated with any of the markers of hyperglycemia in whites independently or in a meta-analysis across ancestries (Tables 1 and 2). This SNP was not available in the CARDIA data set; however, a proxy SNP in perfect LD (r 2 = 1, rs35256014, MAF = 0.06) was present but did not replicate the association with fructosamine in blacks (P = 0.57), and meta-analysis results were also nonsignificant (Table 1 and Supplementary Fig. 13).
An intronic variant, rs59443763, in PRKCA on chromosome 17 (MAF = 0.06), was significantly associated with 5.4% higher percent glycated albumin per minor allele in blacks (P = 4.9 × 10−9, variance explained = 2%) (Table 1 and Supplementary Figs. 14 and 15). It was also associated with fructosamine (P = 9.4 × 10−7) and total glycated albumin (P = 5.8 × 10−7), although these associations did not meet genome-wide significance (Table 2). This SNP was not significant among whites or in trans-ancestry meta-analysis (Tables 1 and 2), but there was limited power to replicate (Supplementary Table 2). No proxy SNPs with r2 > 0.7 were available in the CARDIA data set, and thus replication was not possible for this association.
Sensitivity Analyses
In analyses that excluded participants with undiagnosed diabetes, genome-wide significant results remained for the white sample (N = 7,229) but were no longer present among the reduced sample of black participants (N = 1,878) (Supplementary Table 3).
Of the SNPs significantly associated with fructosamine or glycated albumin, only rs2438321 in blacks (P = 0.002) was significantly associated with serum albumin with a Bonferroni corrected P value (0.05/(4 SNPs * 2 races) = 0.006) (Supplementary Table 4).
Controlling for fasting glucose or HbA1c variants did not reveal any additional genome-wide significant variants, but for glycated albumin, controlling for fasting glucose score attenuated the P value for rs1260326 (P = 0.002) among whites.
Fructosamine, percent glycated albumin, and total glycated albumin had strong, statistically significant genetic correlations (0.92 to 1.17) indicating a large proportion of shared genetics (Supplementary Table 5). Correlations between fasting glucose and HbA1c with the other biomarkers were moderate to substantial but were not significant.
Candidate SNP Analysis
We investigated SNPs previously identified in fasting glucose and HbA1c GWAS for association with fructosamine and percent glycated albumin. Nineteen of the 41 fasting glucose SNPs were nominally (P < 0.05) associated with fasting glucose, and 13 of these associations were in the same direction in blacks and whites (Table 3 and Supplementary Table 6) and 33 had consistent direction with the discovery cohort in whites. Four variants (10%) were study-wide significantly associated with fructosamine and percent glycated albumin in whites, three of which were associated with percent glycated albumin and one of which was associated with fructosamine. No variants were study-wide significantly associated with fructosamine or percent glycated albumin in blacks.
Table 3.
Significance of associations between fasting glucose known genetic determinants and fructosamine and percent glycated albumin in ARIC
| SNP | Chr:position | Closest gene | A1/A2 | Whites |
Blacks |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 freq | P value fructosamine | P value glycated albumin | P value fasting glucose | A1 freq | P value fructosamine | P value glycated albumin | P value fasting glucose | ||||
| rs10747083 |
12:133041618 |
P2RX2 |
G/A |
0.33 |
0.95 |
0.76 |
0.14 |
0.16 |
0.17 |
0.11 |
0.95 |
| rs10830963 |
11:92708710 |
MTNR1B |
G/C |
0.28 |
0.001 |
0.003 |
3.3E-07 |
0.07 |
0.13 |
0.03 |
0.38 |
| rs10885122 |
10:113042093 |
ADRA2A |
T/G |
0.12 |
0.82 |
0.75 |
0.76 |
0.68 |
0.24 |
0.72 |
0.21 |
| rs11071657 |
15:62433962 |
C2CD4B |
G/A |
0.38 |
0.50 |
0.81 |
0.52 |
0.12 |
0.98 |
0.95 |
0.43 |
| rs11558471 |
8:118185733 |
SLC30A8 |
G/A |
0.32 |
0.005 |
8.2E-05 |
1.8E-05 |
0.09 |
0.31 |
0.18 |
0.65 |
| rs11603334 |
11:72432985 |
ARAP1 |
A/G |
0.16 |
0.01 |
0.08 |
0.47 |
0.06 |
0.79 |
0.95 |
0.76 |
| rs11605924 |
11:45873091 |
CRY2 |
A/C |
0.47 |
0.36 |
0.58 |
0.66 |
0.84 |
0.58 |
0.72 |
0.44 |
| rs11708067 |
3:123065778 |
ADCY5 |
G/A |
0.23 |
0.07 |
0.001 |
0.01 |
0.16 |
0.002 |
0.003 |
0.03 |
| rs11715915 |
3:49455330 |
AMT |
T/C |
0.30 |
0.30 |
0.74 |
0.04 |
0.23 |
0.06 |
0.37 |
0.31 |
| rs11920090 |
3:170717521 |
SLC2A2 |
A/T |
0.13 |
0.004 |
0.002 |
0.34 |
0.35 |
0.47 |
0.30 |
0.28 |
| rs13179048 |
5:95542726 |
PCSK1 |
A/C |
0.31 |
0.11 |
0.13 |
0.51 |
0.08 |
0.18 |
0.52 |
0.78 |
| rs1371614 |
2:27152874 |
DPYSL5 |
T/C |
0.25 |
0.47 |
0.78 |
0.69 |
0.36 |
0.18 |
0.11 |
0.21 |
| rs143399767 |
9:96182703 |
YRNA |
C/A |
0.01 |
0.65 |
0.24 |
0.55 |
||||
| rs1483121 |
11:48333360 |
OR4S1 |
A/G |
0.13 |
0.41 |
0.36 |
0.74 |
0.04 |
0.46 |
0.42 |
0.94 |
| rs16913693 |
9:111680359 |
IKBKAP |
G/T |
0.03 |
0.60 |
0.78 |
0.11 |
0.25 |
0.73 |
0.68 |
0.91 |
| rs174550 |
11:61571478 |
FADS1 |
C/T |
0.33 |
0.16 |
0.28 |
0.54 |
0.08 |
0.37 |
0.15 |
0.93 |
| rs17762454 |
6:7213200 |
RREB1 |
T/C |
0.26 |
0.13 |
0.32 |
0.001 |
0.16 |
0.16 |
0.76 |
0.75 |
| rs2191349 |
7:15064309 |
TMEM195, DGKB |
G/T |
0.46 |
0.17 |
0.36 |
0.02 |
0.44 |
0.58 |
0.73 |
0.004 |
| rs2293941 |
13:28491198 |
PDX1 |
A/G |
0.22 |
0.02 |
0.005 |
0.03 |
0.17 |
0.38 |
0.60 |
0.77 |
| rs2302593 |
19:46196634 |
GIPR |
G/C |
0.50 |
1.00 |
0.40 |
0.02 |
0.30 |
0.64 |
0.94 |
0.97 |
| rs2657879 |
12:56865338 |
GLS2 |
G/A |
0.18 |
0.93 |
0.94 |
0.31 |
0.06 |
0.05 |
0.23 |
0.04 |
| rs2722425 |
8:40484239 |
ZMAT4 |
T/C |
0.12 |
0.01 |
0.01 |
0.78 |
0.37 |
0.25 |
0.35 |
0.41 |
| rs340874 |
1:214159256 |
PROX1 |
T/C |
0.45 |
0.42 |
0.82 |
0.40 |
0.17 |
0.36 |
0.74 |
0.15 |
| rs35767 |
12:102875569 |
IGF1 |
G/A |
0.16 |
0.76 |
0.54 |
0.62 |
0.55 |
0.43 |
0.10 |
0.02 |
| rs3736594 |
2:27995781 |
MRPL33 |
C/A |
0.26 |
0.11 |
0.07 |
0.25 |
0.43 |
0.07 |
0.03 |
0.62 |
| rs3783347 |
14:100839261 |
WARS |
T/G |
0.22 |
0.45 |
0.51 |
0.57 |
0.06 |
0.44 |
0.60 |
0.94 |
| rs3829109 |
9:139256766 |
DNLZ |
A/G |
0.29 |
0.003 |
0.13 |
0.16 |
0.18 |
0.51 |
0.39 |
0.46 |
| rs4506565 |
10:114756041 |
TCF7L2 |
T/A |
0.31 |
3.9E-04 |
2.5E-05 |
2.7E-05 |
0.44 |
0.09 |
0.40 |
0.83 |
| rs4607517 |
7:44235668 |
GCK |
A/G |
0.17 |
2.0E-04 |
0.003 |
5.0E-04 |
0.11 |
0.09 |
0.10 |
0.08 |
| rs4841132 |
8:9183596 |
PPP1R3B |
A/G |
0.09 |
0.33 |
0.32 |
0.05 |
0.13 |
0.74 |
0.29 |
0.05 |
| rs560887 |
2:169763148 |
G6PC2 |
T/C |
0.30 |
0.08 |
0.003 |
7.3E-05 |
0.05 |
0.81 |
0.58 |
0.91 |
| rs576674 |
13:33554302 |
KL |
G/A |
0.16 |
0.45 |
0.50 |
0.33 |
0.61 |
0.17 |
0.26 |
0.11 |
| rs6048205 |
20:22559601 |
FOXA2 |
G/A |
0.05 |
0.79 |
0.95 |
0.07 |
0.18 |
0.02 |
0.25 |
0.05 |
| rs6072275 |
20:39743905 |
TOP1 |
A/G |
0.16 |
0.57 |
0.30 |
0.01 |
0.08 |
0.13 |
0.12 |
0.08 |
| rs6943153 |
7:50791579 |
GRB10 |
T/C |
0.31 |
0.67 |
0.46 |
0.16 |
0.70 |
0.09 |
0.77 |
0.61 |
| rs7034200 |
9:4289050 |
GLIS3 |
A/C |
0.49 |
0.19 |
0.04 |
0.03 |
0.61 |
0.18 |
0.02 |
0.11 |
| rs7651090 |
3:185513392 |
IGF2BP2 |
G/A |
0.31 |
0.01 |
0.03 |
0.003 |
0.56 |
0.32 |
0.58 |
0.40 |
| rs7708285 |
5:76425867 |
ZBED3 |
G/A |
0.30 |
0.32 |
0.64 |
0.71 |
0.15 |
0.56 |
0.52 |
0.16 |
| rs780094 |
2:27741237 |
GCKR |
T/C |
0.40 |
0.05 |
5.7E-05 |
1.0E-04 |
0.18 |
0.75 |
0.96 |
0.90 |
| rs7944584 |
11:47336320 |
MADD |
T/A |
0.27 |
0.56 |
0.76 |
0.05 |
0.04 |
0.98 |
0.38 |
0.77 |
| rs9368222 | 6:20686996 | CDKAL1 | A/C | 0.27 | 0.19 | 0.09 | 0.27 | 0.20 | 0.96 | 0.80 | 0.68 |
freq, frequency. Candidate SNPs selected from NHGRI database based on previous genome-wide significant associations. P values that reach study-wide significance for fructosamine and glycated albumin, P < 3.0 × 10−4 (0.05/(2*2*41)), and P values that reach nominal significance (P < 0.05) for fasting glucose are in boldface type.
Thirty-one of 46 previously identified HbA1c SNPs were nominally associated with HbA1c, 15 of which were associated in the same direction in blacks and whites (Table 4 and Supplementary Table 7), and 43 had consistent direction with the discovery cohort in whites. Five SNPs (11%) demonstrated a study-wide significant association with fructosamine or glycated albumin in whites. All variants associated with multiple glycemic biomarkers had effects in the same direction.
Table 4.
Significance of associations between HbA1c known genetic determinants and fructosamine and percent glycated albumin in ARIC
| SNP | Chr:position | Closest gene | A1/A2 | Whites |
Blacks |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 freq | P value fructosamine | P value glycated albumin | P value HbA1c | A1 freq | P value fructosamine | P value glycated albumin | P value HbA1c | ||||
| rs1046896 |
17:80685533 |
FN3KRP |
T/C |
0.31 |
0.034 |
0.048 |
0.052 |
0.24 |
0.661 |
0.455 |
0.585 |
| rs10774625 |
12:111910219 |
ATXN2 |
G/A |
0.48 |
0.571 |
0.032 |
0.008 |
0.09 |
0.221 |
0.142 |
0.503 |
| rs10823343 |
10:71091013 |
HK1 |
G/A |
0.26 |
0.452 |
0.065 |
1.1E-06 |
0.45 |
0.808 |
0.529 |
0.899 |
| rs10830963 |
11:92708710 |
MTNR1B |
G/C |
0.28 |
0.001 |
0.003 |
0.006 |
0.07 |
0.134 |
0.035 |
0.222 |
| rs11248914 |
16:293562 |
ITFG3 |
C/T |
0.35 |
0.011 |
0.229 |
0.055 |
0.36 |
0.067 |
0.354 |
0.485 |
| rs11558471 |
8:118185733 |
SLC30A8 |
G/A |
0.32 |
0.005 |
8.2E-05 |
0.010 |
0.09 |
0.309 |
0.176 |
0.989 |
| rs11603334 |
11:72432985 |
ARAP1 |
A/G |
0.16 |
0.005 |
0.078 |
0.228 |
0.06 |
0.788 |
0.950 |
0.734 |
| rs11708067 |
3:123065778 |
ADCY5 |
G/A |
0.23 |
0.070 |
0.001 |
0.034 |
0.16 |
0.002 |
0.003 |
0.074 |
| rs11954649 |
5:157055491 |
SOX30 |
G/C |
0.00 |
NA |
NA |
NA |
0.05 |
0.812 |
0.220 |
0.184 |
| rs11964178 |
6:109562035 |
C6orf183 |
G/A |
0.43 |
0.861 |
0.844 |
0.702 |
0.36 |
0.180 |
0.748 |
0.475 |
| rs12621844 |
2:48414735 |
FOXN2 |
C/T |
0.39 |
0.732 |
0.294 |
0.358 |
0.84 |
0.829 |
0.853 |
0.666 |
| rs12819124 |
12:48409054 |
RP1 |
A/C |
0.47 |
0.765 |
0.930 |
0.001 |
0.21 |
0.500 |
0.826 |
0.146 |
| rs13134327 |
4:144659795 |
FREM3 |
A/G |
0.32 |
0.800 |
0.662 |
0.031 |
0.30 |
0.220 |
0.854 |
0.772 |
| rs1402837 |
2:169757354 |
G6PC2 |
T/C |
0.22 |
0.016 |
9.3E-05 |
2.7E-07 |
0.31 |
0.471 |
0.728 |
0.656 |
| rs1558902 |
16:53803574 |
FTO |
A/T |
0.41 |
0.545 |
0.017 |
0.131 |
0.11 |
0.065 |
0.176 |
0.514 |
| rs17509001 |
2:24021231 |
ATAD2B |
C/T |
0.14 |
0.309 |
0.152 |
0.027 |
0.09 |
0.604 |
0.524 |
0.802 |
| rs17533903 |
19:17256523 |
MYO9B |
A/G |
0.21 |
0.577 |
0.167 |
5.0E-04 |
0.24 |
0.403 |
0.232 |
0.689 |
| rs17747324 |
10:114752503 |
TCF7L2 |
C/T |
0.23 |
1.7E-04 |
3.2E-05 |
1.8E-04 |
0.07 |
0.396 |
0.755 |
0.890 |
| rs1800562 |
6:26093141 |
HFE |
A/G |
0.06 |
0.337 |
0.128 |
0.001 |
0.01 |
0.846 |
0.670 |
0.146 |
| rs198846 |
6:26107463 |
HFE |
A/G |
0.16 |
0.390 |
0.198 |
0.123 |
0.88 |
0.533 |
0.270 |
0.260 |
| rs2110073 |
12:7075882 |
PHB2 |
T/C |
0.10 |
0.321 |
0.938 |
0.002 |
0.42 |
0.177 |
0.349 |
0.160 |
| rs2383208 |
9:22132076 |
MTAP |
G/A |
0.18 |
0.008 |
0.019 |
0.007 |
0.19 |
0.293 |
0.763 |
0.596 |
| rs2408955 |
12:48499131 |
SENP1 |
G/T |
0.48 |
0.567 |
0.879 |
0.002 |
0.63 |
0.813 |
0.824 |
0.632 |
| rs267738 |
1:150940625 |
CERS2 |
G/T |
0.21 |
0.444 |
0.434 |
0.511 |
0.04 |
0.853 |
0.745 |
0.297 |
| rs2779116 |
1:158585415 |
SPTA1 |
T/C |
0.27 |
0.714 |
0.792 |
9.9E-06 |
0.22 |
0.111 |
0.167 |
0.961 |
| rs282587 |
13:113351662 |
ATP11A |
G/A |
0.12 |
0.586 |
0.248 |
1.0E-04 |
0.69 |
0.050 |
0.142 |
0.246 |
| rs3824065 |
7:44247258 |
GCK |
T/C |
0.42 |
3.3E-04 |
0.002 |
0.125 |
0.24 |
0.086 |
0.134 |
0.910 |
| rs4607517 |
7:44235668 |
GCK |
A/G |
0.17 |
2.0E-04 |
0.003 |
5.7E-05 |
0.11 |
0.088 |
0.097 |
0.244 |
| rs4737009 |
8:41630405 |
ANK1 |
A/G |
0.24 |
0.126 |
0.515 |
0.018 |
0.46 |
0.274 |
0.461 |
0.361 |
| rs4745982 |
10:71089843 |
HK1 |
G/T |
0.07 |
0.190 |
0.263 |
5.8E-06 |
0.10 |
0.889 |
0.921 |
0.602 |
| rs4820268 |
22:37469591 |
TMPRSS6 |
G/A |
0.46 |
0.264 |
0.350 |
0.010 |
0.72 |
0.073 |
0.029 |
0.181 |
| rs560887 |
2:169763148 |
G6PC2 |
T/C |
0.30 |
0.080 |
0.003 |
0.003 |
0.95 |
0.811 |
0.583 |
0.861 |
| rs579459 |
9:136154168 |
ABO |
C/T |
0.23 |
0.268 |
0.340 |
0.001 |
0.13 |
0.210 |
0.487 |
0.600 |
| rs592423 |
6:139840693 |
CITED2 |
A/C |
0.46 |
0.514 |
0.535 |
0.063 |
0.61 |
0.323 |
0.605 |
0.333 |
| rs6474359 |
8:41549194 |
ANK1 |
C/T |
0.03 |
0.979 |
0.486 |
4.3E-04 |
0.27 |
0.077 |
0.047 |
0.879 |
| rs6980507 |
8:42383084 |
SLC20A2 |
A/G |
0.39 |
0.256 |
0.873 |
0.015 |
0.48 |
0.514 |
0.557 |
0.663 |
| rs7040409 |
9:91503236 |
C9orf47 |
G/C |
0.07 |
0.499 |
0.765 |
0.001 |
0.26 |
0.028 |
0.005 |
0.115 |
| rs7616006 |
3:12267648 |
SYN2 |
G/A |
0.43 |
0.705 |
0.385 |
0.181 |
0.36 |
0.974 |
0.959 |
0.836 |
| rs761772 |
17:76122078 |
TMC6 |
C/T |
0.13 |
0.077 |
0.109 |
0.009 |
0.13 |
0.480 |
0.361 |
0.861 |
| rs7756992 |
6:20679709 |
CDKAL1 |
G/A |
0.27 |
0.180 |
0.094 |
0.292 |
0.42 |
0.856 |
0.997 |
0.995 |
| rs8192675 |
3:170724883 |
SLC2A2 |
C/T |
0.30 |
5.4E-05 |
6.1E-05 |
0.010 |
0.29 |
0.232 |
0.239 |
0.024 |
| rs837763 |
16:88853729 |
CDT1 |
C/T |
0.44 |
0.591 |
0.804 |
0.046 |
0.56 |
0.492 |
0.554 |
0.533 |
| rs857691 |
1:158626378 |
SPTA1 |
T/C |
0.25 |
0.803 |
0.492 |
5.6E-06 |
0.30 |
0.092 |
0.226 |
0.894 |
| rs9604573 |
13:114542858 |
GAS6 |
A/G |
0.26 |
0.482 |
0.300 |
0.687 |
0.24 |
0.902 |
0.833 |
0.425 |
| rs9818758 |
3:49382925 |
USP4 |
A/G |
0.17 |
0.240 |
0.870 |
0.780 |
0.09 |
0.297 |
0.297 |
0.642 |
| rs9914988 | 17:27183104 | ERAL1 | G/A | 0.21 | 0.307 | 0.993 | 0.058 | 0.66 | 0.252 | 0.080 | 0.342 |
freq, frequency. Candidate SNPs selected from NHGRI database based on previous genome-wide significant associations. P values that reach study-wide significance for fructosamine and glycated albumin, P < 2.7 × 10−4 (0.05/(2*2*46)), and P values that reach nominal significance (P < 0.05) for HbA1c are in boldface type.
Percent Variance Explained
SNPs associated with fasting glucose (N = 41) (Table 3) explained 1.4% of the variance in fructosamine, 3.2% of the variance in percent glycated albumin, and 1.9% of the variance in total glycated albumin among whites. Taking SNPs associated with serum albumin from the GWAS catalog explained 0.4% of the variance of fructosamine, 1.1% of the variance of percent glycated albumin, and 0.7% of the variance of total glycated albumin among the white sample.
Discussion
We identified four SNPs significantly associated with fructosamine and glycated albumin among either whites or blacks, one which replicated in a second cohort and three not previously associated with glycemic traits. Several known fasting glucose and HbA1c SNPs were significantly associated with fructosamine or glycated albumin.
Among whites, rs1260326 was associated with percent glycated albumin. This variant reflects the same signal associated with type 2 diabetes and fasting glucose: it is in perfect LD with a known type 2 diabetes variant (r2 = 1 among 1000 Genomes phase 3 Europeans with rs145819220, from a recent large type 2 diabetes GWAS [38]) and in strong LD with a known fasting glucose variant (r2 = 0.91 with rs780094 [37,38]). rs1260326 is located in glucokinase (hexokinase 4) regulator (GCKR), which encodes a regulatory protein primarily active in the liver that inhibits glucokinase (GCK), the enzyme in the first step of glycolysis and involved in converting glucose to glycogen for storage. GCK is considered a glucose sensor that helps maintain glucose homeostasis. The GCKR protein product inhibits the activity of GCK, increasing serum glucose levels. GCKR is an established type 2 diabetes gene (39–41) and is associated with multiple other traits including kidney disease, triglyceride levels, and Crohn disease (42–44). Thus, this variant likely represents part of a glycemic pathway, but it is interesting that in our study it is only significantly associated with one measure of hyperglycemia, approached significance with fasting glucose (although controlling for fasting glucose score made this variant nonsignificant) and total glycated albumin, but is not associated with fructosamine or HbA1c, given the moderate to strong correlations and genetic correlations among the biomarkers (Supplementary Tables 5 and 8). That GCKR is primarily expressed in the liver rather than the pancreas (45,46) aligns with the finding of association with fasting glucose, which measures hepatic glucose output. Albumin is also produced by the liver, while erythrocytes and hemoglobin are not likely affected by liver function, thus perhaps hepatic-specific genetic factors would be more likely to associate with percent glycated albumin levels than with HbA1c. It is also possible that HbA1c, affected by other glucose-altering factors, may mask the effect of rs1260326 on GCKR. Adjustment for serum albumin may explain the association with percent glycated albumin but not fructosamine.
We also identified several variants of potential interest that were significant in ARIC but lacked replication. Among whites, rs34459162, in RCN3, was associated with fructosamine and total glycated albumin. RCN3 encodes reticulocalbin 3, an EF-hand calcium binding domain (47). This SNP was not associated with serum albumin in our analysis, but a SNP in perfect LD with rs34459162, rs2280401, was associated with total protein in a Japanese population (48) and serum albumin in an East Asian population (49), indicating a possible impact on fructosamine and glycated albumin through nonglycemic pathways. Among blacks, we found two novel variants: rs2438321 (intergenic and closest to CNTN5, which encodes a glycosylphosphatidylinositol-anchored neuronal membrane protein, a member of the immunoglobulin superfamily and the contactin family) associated with fructosamine and rs59443763 (PRKCA, which encodes protein kinase C alpha, ubiquitous in cellular processes) associated with percent glycated albumin at a genome-wide level of significance. There is no prior literature on either of these as potential glycemic loci in diabetes. While we had sufficient power to replicate the results for whites in CARDIA (Supplementary Table 2), we had low power among blacks, which may be why these SNPs did not replicate. These variants became nonsignificant after excluding undiagnosed diabetes, which may be due to the greater number of individuals with undiagnosed diabetes among blacks than whites. Blacks had higher values of glycemic biomarkers, thus removing case subjects with undiagnosed diabetes could have had a greater impact on associations among blacks than whites. These variants should be evaluated in larger African ancestry data sets as they become available. rs34459162, rs2438321, and rs59443763 are of potential interest, but as these SNPs currently lack replication, we cannot rule out false positive results.
Results varied by ancestry for the SNPs available in both blacks and whites: neither SNP was significant in both blacks and whites, and meta-analyses results were nonsignificant. While this may partially be explained by differing allele frequencies (rs1260326: 0.41 in whites, 0.14 in blacks; rs2438321: 0.24 in whites, 0.11 in blacks), a differential effect by ancestry on fructosamine and glycated albumin is also possible. This may be particularly true for rs2438321, where the direction of effect differs across ancestries.
In addition to investigating fructosamine and glycated albumin individually, comparing to traditional glycemic markers (fasting glucose and HbA1c) can help to clarify the biological pathways involved in diabetes. Fasting glucose–related SNPs explained almost twice the variance of percent glycated albumin than that of fructosamine. This may reflect the adjustment for serum albumin with percent glycated albumin and not with fructosamine, allowing percent glycated albumin levels to be influenced more by glucose levels and less by albumin levels. However, albumin SNPs also explained more variance of percent glycated albumin than that of fructosamine or total glycated albumin. Given the small percentages, it is difficult to draw first conclusions from these results.
Only five HbA1c variants were significantly associated with fructosamine or glycated albumin. This is consistent with the findings that the majority of HbA1c variants are related to erythrocyte and hemoglobin factors that we would not expect to be related to fructosamine or glycated albumin. Many associations of fructosamine or glycated albumin with HbA1c SNPs or fasting glucose SNPs were present in whites but not blacks. This is not surprising given that the SNPs were originally detected in whites and that our sample size was larger for whites, with corresponding higher power to detect moderate associations. Not all of the previously discovered SNPs for fasting glucose and HbA1c replicated for those outcomes in our sample, but this again may have to do with lack of power.
We found that both glycemic and nonglycemic genetic factors influenced fructosamine and glycated albumin levels. We identified a likely glycemic variant in a gene associated with type 2 diabetes (GCKR), supporting its role in diabetes biology, and a likely nonglycemic variant in a gene (RCN3) that may reflect the biology of a biomarker (i.e., influencing amount of serum protein available to be glycated) rather than the biology of type 2 diabetes. This contribution of glycemic and nonglycemic variants is similar to the pattern of genetic contribution to HbA1c, for which the majority of genetic variants are nonglycemic (18,19). In our study, previously identified nonglycemic variants (18–20) explained 3.4% of the variance in HbA1c and the glycemic variants explained 2.1% (Supplementary Table 9). Despite the previous studies having much larger sample sizes (and thus more power to detect associations with HbA1c), the percent variance explained we found for fructosamine (0.6% by likely nonglycemic rs34459162) and glycated albumin (0.3% by likely glycemic rs1260326) was of a similar magnitude. Both Soranzo et al. (18) and Chen et al. (19) found that taking nonglycemic variants into account modestly impacted diabetes reclassification, and Wheeler et al. (20) found a more substantial effect. Given that that future larger studies on fructosamine and glycated albumin will likely reveal other significant variants, it will be important to determine whether the effect of nonglycemic variants is substantial enough to impact the clinical interpretation of fructosamine and glycated albumin.
A major limitation of this study was the limited sample size, particularly the smaller sample size in blacks. The differences in ancestries make replication of results difficult, particularly if allele frequencies differ, and warrant more studies focused on multiethnic populations. Also, the lack of an available SNP or proxy for rs59443763 in CARDIA, possibly due to the imputation reference panel (HapMap Phase II), impeded our ability to evaluate replication of this finding. In addition, the sample size for our replication cohort was much smaller than our discovery cohort, limiting our power to replicate the significant ARIC findings in blacks.
In summary, through GWAS in a community-based population of blacks and whites, we identified and replicated two significant variants associated with fructosamine and/or glycated albumin, one of which was novel. These variants map into a likely glycemic, known diabetes gene and a likely nonglycemic gene. This highlights the utility of examining genetics of diabetes biomarkers both for providing insight into the pathophysiology of diabetes and for better understanding glucose-independent influences on measures of hyperglycemia.
Supplementary Material
Article Information
Acknowledgments. This article is dedicated to our dear friend and colleague, Dr. W.H. Linda Kao (1972–2014). We would like to acknowledge Linda’s contributions to the design of the study and early discussions of this work. The authors thank the staff and participants of the ARIC Study for their important contributions.
Funding. The ARIC Study has been funded in whole or in part with federal funds from the National Heart, Lung, and Blood Institute (NHLBI), National Institutes of Health (NIH), Department of Health and Human Services (contract numbers HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I, and HHSN268201700005I and grant numbers R01HL087641 and R01HL086694), NHGRI contract U01HG004402, and NIH contract HHSN268200625226C. Infrastructure was partly supported by grant number UL1RR025005, a component of the NIH and NIH Roadmap for Medical Research. Reagents for the glycated albumin assays were donated by the Asahi Kasei Corporation. Reagents for the fructosamine assays were donated by Roche Diagnostics Corporation. S.J.L. was supported by an institutional training grant from the NIH/NHLBI (T32 HL007024). E.S. was supported by NIH National Institute for Diabetes and Digestive and Kidney Diseases (NIDDK) grants K24DK106414 and R01DK089174. A.K. was supported German Research Foundation, Deutsche Forschungsgemeinschaft (KO 3598/3-1). The CARDIA study is conducted and supported by the NHLBI in collaboration with the University of Alabama at Birmingham (HHSN268201300025C and HHSN268201300026C), Northwestern University (HHSN268201300027C), University of Minnesota (HHSN268201300028C), Kaiser Foundation Research Institute (HHSN268201300029C), and The Johns Hopkins University School of Medicine (HHSN268200900041C). CARDIA is also partially supported by the Intramural Research Program of the National Institute on Aging (NIA) and an intra-agency agreement between NIA and NHLBI (AG0005). Genotyping was funded as part of the NHLBI Candidate Gene Association Resource (N01-HC-65226) and the NHGRI Gene Environment Association Studies (GENEVA) (U01-HG004729, U01-HG04424, and U01-HG004446). J.C. was supported by NIH NIDDK grants K01DK095928 and P30DK079626 and NIH National Center for Advancing Translational Sciences grant UL1TR000165.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. S.J.L. and N.M.M. wrote the manuscript. M.L. and A.S.B. performed data analysis and reviewed and edited the manuscript. K.E.N., H.M., A.M., A.P.C., J.S.P., E.B., R.S., L.J.R.-T., and J.C. reviewed and edited the manuscript. P.D., A.K., and E.S. contributed to discussion, provided guidance, and reviewed and edited the manuscript. S.J.L. and M.L. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. An earlier version of this work was presented as a poster at the 75th Scientific Sessions of the American Diabetes Association, Boston, MA, 5–9 June 2015.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db17-1362/-/DC1.
References
- 1.American Diabetes Association. Standards of Medical Care in Diabetes—2017. Diabetes Care 2017;40(Suppl. 1):S1–S142 [DOI] [PubMed]
- 2.Sacks DB. A1C versus glucose testing: a comparison. Diabetes Care 2011;34:518–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selvin E, Rawlings AM, Grams M, et al. . Fructosamine and glycated albumin for risk stratification and prediction of incident diabetes and microvascular complications: a prospective cohort analysis of the Atherosclerosis Risk in Communities (ARIC) Study. Lancet Diabetes Endocrinol 2014;2:279–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selvin E, Rawlings AM, Lutsey PL, et al. . Fructosamine and glycated albumin and the risk of cardiovascular outcomes and death. Circulation 2015;132:269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johns Hopkins Medicine. POC-IT Guides: alternative markers of glycemia: fructosamine, glycated albumin, 1,5-AG [Internet]. Available from http://www.hopkinsguides.com/hopkins/ub/view/Johns_Hopkins_Diabetes_Guide/547055/all/Alternative_markers_of_glycemia:_fructosamine__glycated_albumin__1_5_AG. Accessed 20 January 2018
- 6.Joslin Diabetes Center. Home blood glucose (sugar) monitoring, hemoglobin A1c testing, and fructosamine tests [Internet]. Available from http://www.joslin.org/info/home_blood_glucose_sugar_monitoring_hemoglobin_a1c_testing_and_fructosamine_tests.html. Accessed 20 January 2018
- 7.He X, Ying L, Ma X, et al. . An additional measurement of glycated albumin can help prevent missed diagnosis of diabetes in Chinese population. Clin Chim Acta 2017;475:188–192 [DOI] [PubMed] [Google Scholar]
- 8.Malmström H, Walldius G, Grill V, Jungner I, Gudbjörnsdottir S, Hammar N. Fructosamine is a useful indicator of hyperglycaemia and glucose control in clinical and epidemiological studies--cross-sectional and longitudinal experience from the AMORIS cohort. PLoS One 2014;9:e111463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welsh KJ, Kirkman MS, Sacks DB. Role of glycated proteins in the diagnosis and management of diabetes: research gaps and future directions. Diabetes Care 2016;39:1299–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danese E, Montagnana M, Nouvenne A, Lippi G. Advantages and pitfalls of fructosamine and glycated albumin in the diagnosis and treatment of diabetes. J Diabetes Sci Technol 2015;9:169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koga M, Kasayama S. Clinical impact of glycated albumin as another glycemic control marker. Endocr J 2010;57:751–762 [DOI] [PubMed] [Google Scholar]
- 12.Rubinow KB, Hirsch IB. Reexamining metrics for glucose control. JAMA 2011;305:1132–1133 [DOI] [PubMed] [Google Scholar]
- 13.Cohen RM, Sacks DB. Comparing multiple measures of glycemia: how to transition from biomarker to diagnostic test? Clin Chem 2012;58:1615–1617 [DOI] [PubMed] [Google Scholar]
- 14.Parrinello CM, Selvin E. Beyond HbA1c and glucose: the role of nontraditional glycemic markers in diabetes diagnosis, prognosis, and management. Curr Diab Rep 2014;14:548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armbruster DA. Fructosamine: structure, analysis, and clinical usefulness. Clin Chem 1987;33:2153–2163 [PubMed] [Google Scholar]
- 16.Araki T, Ishikawa Y, Okazaki H, et al.; Japanese Red Cross GA Research Group . Introduction of glycated albumin measurement for all blood donors and the prevalence of a high glycated albumin level in Japan. J Diabetes Investig 2012;3:492–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Human Genome Research Institute. GWAS Catalog: the NHGRI-EBI Catalog of published genome-wide association studies. [Internet]. Available from www.ebi.ac.uk/gwas Accessed 30 June 2017
- 18.Soranzo N, Sanna S, Wheeler E, et al.; WTCCC . Common variants at 10 genomic loci influence hemoglobin A1C levels via glycemic and nonglycemic pathways [published correction appears in Diabetes 60:1050–1051]. Diabetes 2010;59:3229–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen P, Takeuchi F, Lee JY, et al.; CHARGE Hematology Working Group . Multiple nonglycemic genomic loci are newly associated with blood level of glycated hemoglobin in East Asians. Diabetes 2014;63:2551–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler E, Leong A, Liu CT, et al.; EPIC-CVD Consortium; EPIC-InterAct Consortium; Lifelines Cohort Study . Impact of common genetic determinants of hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: a transethnic genome-wide meta-analysis. PLoS Med 2017;14:e1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dupuis J, Langenberg C, Prokopenko I, et al.; DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators . New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 2010;42:105–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scott RA, Lagou V, Welch RP, et al.; DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium . Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet 2012;44:991–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu CT, Raghavan S, Maruthur N, et al.; AAAG Consortium; CARe Consortium; COGENT-BP Consortium; eMERGE Consortium; MEDIA Consortium; MAGIC Consortium . Trans-ethnic meta-analysis and functional annotation illuminates the genetic architecture of fasting glucose and insulin. Am J Hum Genet 2016;99:56–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The ARIC Investigators The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. Am J Epidemiol 1989;129:687–702 [PubMed] [Google Scholar]
- 25.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009;5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–909 [DOI] [PubMed] [Google Scholar]
- 27.Selvin E, Coresh J, Zhu H, Folsom A, Steffes MW. Measurement of HbA1c from stored whole blood samples in the Atherosclerosis Risk in Communities Study. J Diabetes 2010;2:118–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007;39:906–913 [DOI] [PubMed] [Google Scholar]
- 29.Rosner B. Fundamentals of Biostatistics. Duxbury, Thomson-Brooks/Cole, 2006 [Google Scholar]
- 30.Pattaro C, Teumer A, Gorski M, et al.; ICBP Consortium; AGEN Consortium; CARDIOGRAM; CHARGe-Heart Failure Group; ECHOGen Consortium . Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun 2016;7:10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mägi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics 2010;11:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bulik-Sullivan B, Finucane HK, Anttila V, et al.; ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3 . An atlas of genetic correlations across human diseases and traits. Nat Genet 2015;47:1236–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedman GD, Cutter GR, Donahue RP, et al. . CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol 1988;41:1105–1116 [DOI] [PubMed] [Google Scholar]
- 34.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010;34:816–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 2010;11:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnold M, Raffler J, Pfeufer A, Suhre K, Kastenmüller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics 2015;31:1334–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott RA, Scott LJ, Mägi R, et al.; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium . An expanded genome-wide association study of type 2 diabetes in Europeans. Diabetes 2017;66:2888–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bi M, Kao WH, Boerwinkle E, et al. . Association of rs780094 in GCKR with metabolic traits and incident diabetes and cardiovascular disease: the ARIC Study. PLoS One 2010;5:e11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Liu L, Zhao J, et al. . Large scale meta-analyses of fasting plasma glucose raising variants in GCK, GCKR, MTNR1B and G6PC2 and their impacts on type 2 diabetes mellitus risk. PLoS One 2013;8:e67665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohlke KL, Boehnke M. Recent advances in understanding the genetic architecture of type 2 diabetes. Hum Mol Genet 2015;24(R1):R85–R92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Köttgen A, Pattaro C, Böger CA, et al. . New loci associated with kidney function and chronic kidney disease. Nat Genet 2010;42:376–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sparsø T, Andersen G, Nielsen T, et al. . The GCKR rs780094 polymorphism is associated with elevated fasting serum triacylglycerol, reduced fasting and OGTT-related insulinaemia, and reduced risk of type 2 diabetes. Diabetologia 2008;51:70–75 [DOI] [PubMed] [Google Scholar]
- 44.Umeno J, Asano K, Matsushita T, et al. . Meta-analysis of published studies identified eight additional common susceptibility loci for Crohn’s disease and ulcerative colitis. Inflamm Bowel Dis 2011;17:2407–2415 [DOI] [PubMed] [Google Scholar]
- 45.Beer NL, Tribble ND, McCulloch LJ, et al. . The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet 2009;18:4081–4088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honoré B. The rapidly expanding CREC protein family: members, localization, function, and role in disease. BioEssays 2009;31:262–277 [DOI] [PubMed] [Google Scholar]
- 48.Kamatani Y, Matsuda K, Okada Y, et al. . Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet 2010;42:210–215 [DOI] [PubMed] [Google Scholar]
- 49.Kim YJ, Go MJ, Hu C, et al.; MAGIC consortium . Large-scale genome-wide association studies in East Asians identify new genetic loci influencing metabolic traits. Nat Genet 2011;43:990–995 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.