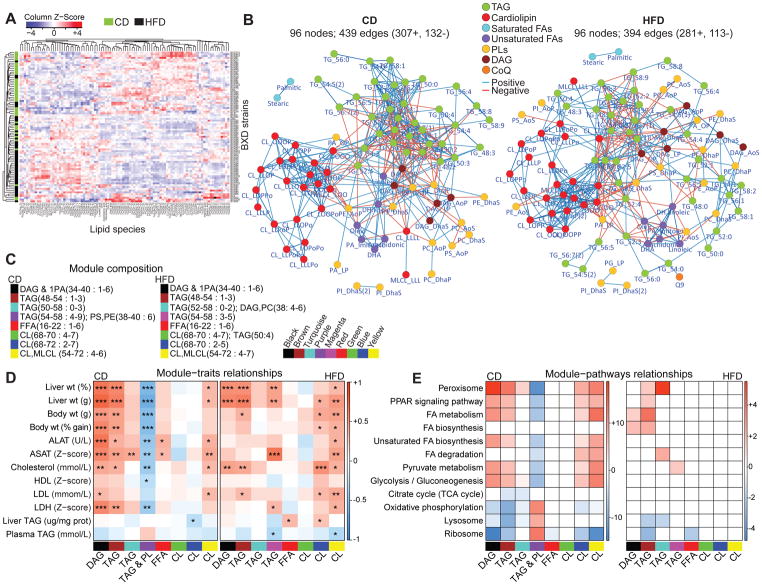
Figure 1. Liver lipid species profile and their association with physiological and molecular traits.
(A) Heatmap analysis with unsupervised hierarchical clustering of 96 lipid species for each BXD cohort shows mixed dietary and genetic impact. (B) Spearman correlation network (p<1e-04) of all lipid species measured in CD and HFD. Lipid species are color coded as 7 major lipid classes. The side chain FA composition of lipids has been abbreviated (O, oleic; P, palmitic; Po, palmitoleic; S, stearic; L, linoleic and Dha, docosahexaenoic acid. Refer to table S1 for abbreviation and composition). (C) Legend of the module composition, indicating the range of total number of carbons and degree of unsaturation. (D) Lipid module-clinical trait correlation. Each cell is color-coded by the Pearson’s correlation coefficient according to the legend color on the right. The stars in the cells represent the p-value of the correlation (p < 0.05 *, p < 0.01 **, p < 0.001 ***). (E) Module and its corresponding KEGG enriched pathway correlation. Red and blue cells represent the enriched pathways with the positively (scale bar: log10 p-value) and negatively (scale bar: -log10 p-value) correlated proteins respectively. Lipid classes hereafter are abbreviated as follows: triacylglycerol, TAG/TG; diacylglycerol, DAG; free-fatty acid, FFA; phospholipid, PL (phosphatidic acid, PA; phosphatidylcholine, PC; phosphatidylethanolamine, PE; phosphatidylglycerol, PG; phosphatidylinositol, PI; phosphatidylserine, PS); cardiolipin, CL; monolysocardiolipin; MLCL. See also Figures S1, S2 and Tables S1 and S2.