Figure 6. Potential interactions involving the three key residues in the BK VSD.
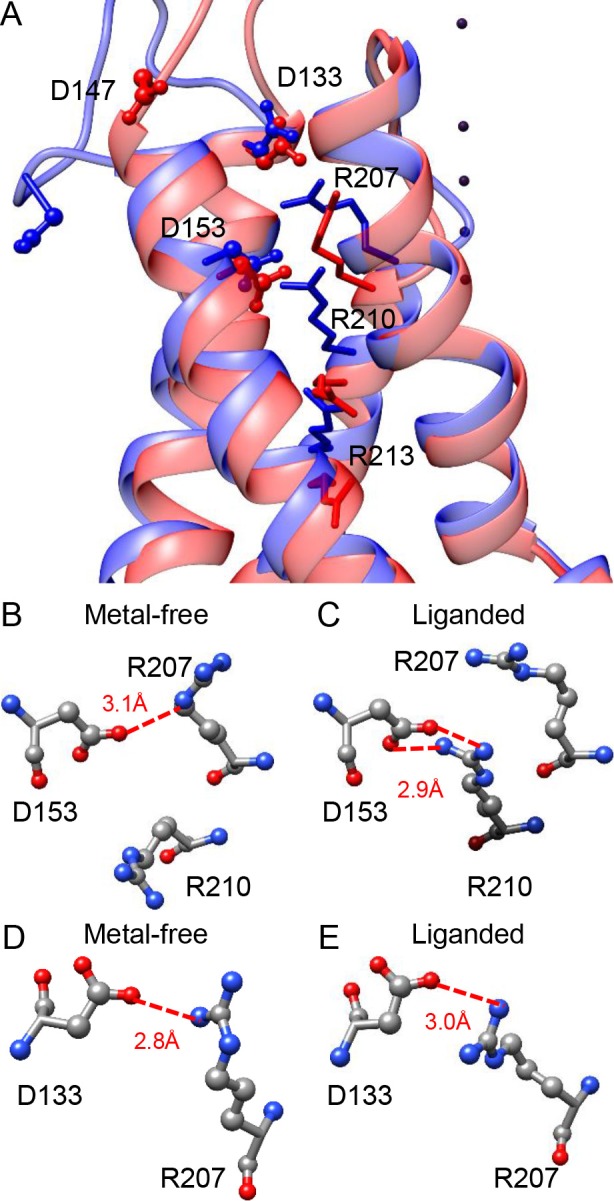
(A) The VSDs of metal-free (red) and liganded (blue) mSlo1 structures with three key acidic residues rendered as ball-and-chain and three conserved S4 arginines (homologous to Shaker R2, R3 and R4) rendered as sticks. The four dark purple dots are K+s in the BK selectivity filter, which are included as a reference for vertical displacement. The two structures are superimposed by the pore-loop. (B) D153 may form a hydrogen bond with R207 in the metal-free mSlo1 structure. The distance between the side chains of D153 and R210 is more than 6 Å, which is too far for hydrogen bonding. (C) D153 may form a hydrogen bond with R210 in the liganded mSlo1 structure. The distance between the side chains of D153 and R207 is more than 4 Å, which is also beyond the limit of effective hydrogen bonding. (D–E) The distances between the side chains of D133 and R207 in both the metal-free (D) and the liganded (E) structures are within 3 Å, allowing potential interaction between D133 and R207 in both conformations.
