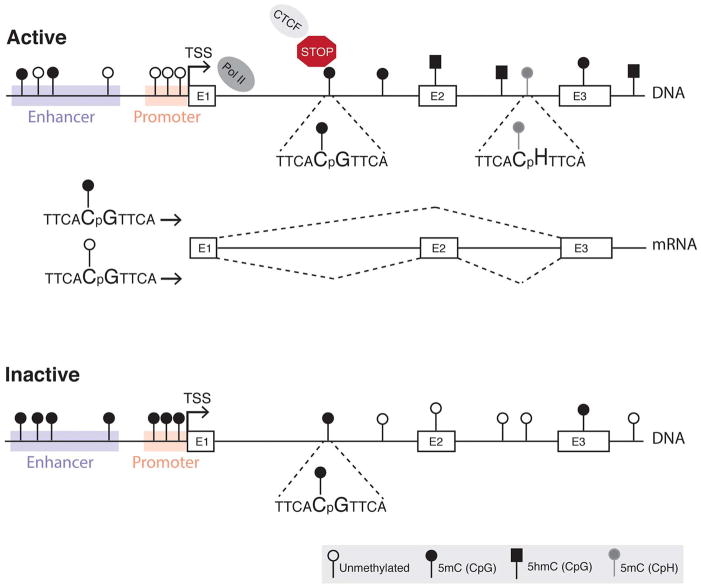
Fig. 1.
Context-dependent roles of DNA methylation. In active genes, DNA methylation is usually absent at the promoter, is low at enhancer regions and variable at gene bodies (Jones, 2012). Neuronal genes may harbor 5hmC and 5mC in the CpG context and/or 5mC in the CpH context (H = A, T or C) at their gene bodies (Lister et al., 2013). CTCF links DNA methylation to RNA splicing by binding to DNA near an alternative exon (E2), halting RNA polymerase and allowing the incorporation of E2 into the transcript (Shukla et al., 2011). DNA methylation can prevent CTCF binding, and thereby the incorporation of alternative exons. Inactive genes usually are methylated at their promoter and enhancer regions, and contain variable methylation at gene bodies with no 5hmC or 5mC in the CpH context.