Fig. 6.
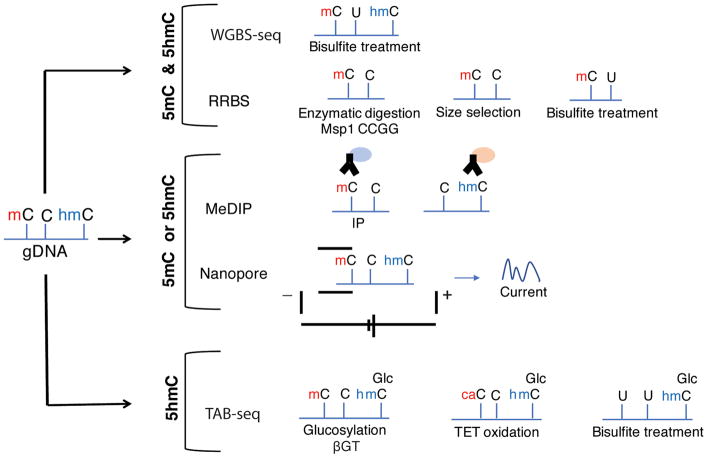
Survey of DNA methylation. Methods to identify 5-methyl-cytosine (5mC) and 5-methyl-hydroxy-cytosine (5hmC) are frequently used in combination with polymerase chain reaction (PCR), next-generation sequencing, or DNA microarrays. Some of these methods, such as genomewide bisulfite sequencing (WGBS-seq) and reduced representation bisulfite sequencing (RRBS), cannot distinguish between 5mC and 5hmC. In bisulfite sequencing, a bisulfite salt converts all unmethylated cytosines to uracyl. In RRBS, specific methylation-insensitive enzymes are used to digest DNA and, after size selection, only a fraction of the genome biased towards CpG rich regions is surveyed (Meissner et al., 2005). To distinguish 5mC from 5hmC, specific antibodies are used in Methylated DNA immunoprecipitation (MeDIP) method (Thu et al., 2009). Nanopore sequencing can detect the electrolytic current signals of base modifications by moving the single-strand DNA molecule through a protein pore (Laszlo et al., 2013). In Tet-assisted bisulfite sequencing (TAB-seq) (Yu et al., 2012), 5hmC is glucosylated by β-glucosyltransferase (βGT) to protect 5hmC from the action of TET enzymes, which convert the 5mC into 5caC. Bisulfite treatment then converts 5caC into U leaving the glucosylated 5hmC intact.