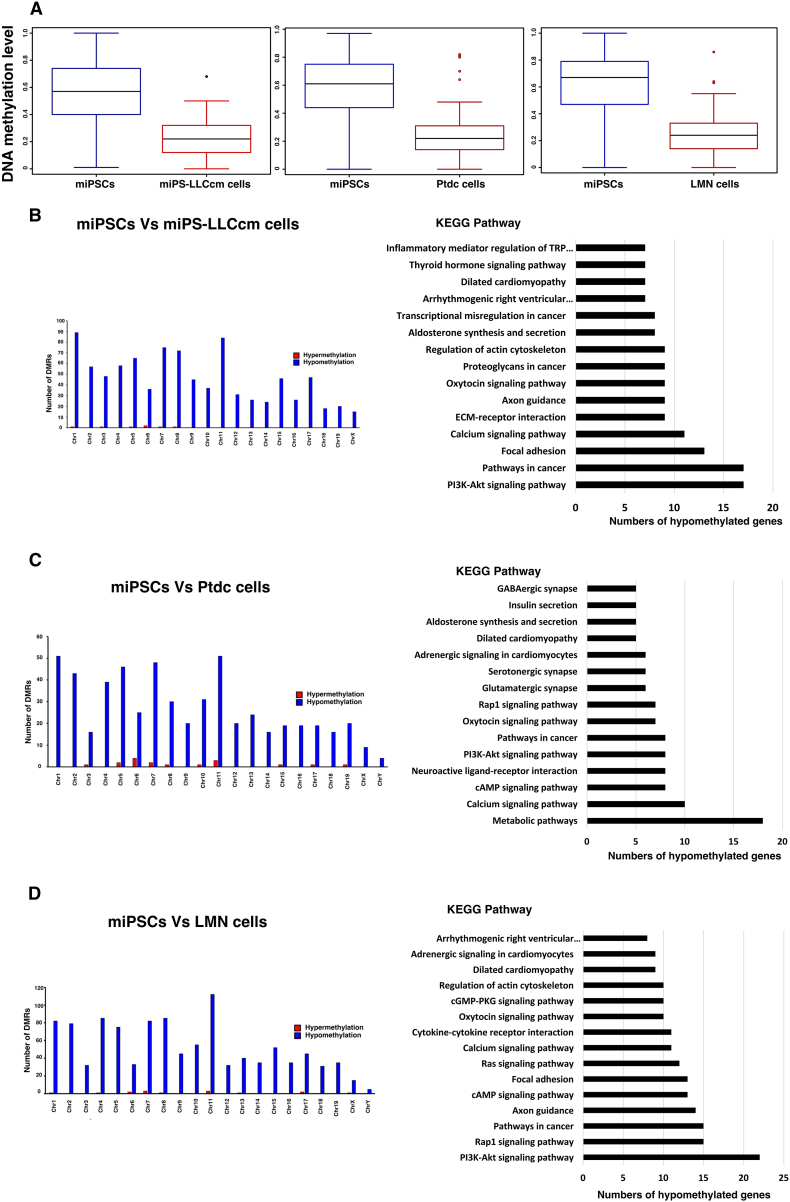
Figure 5.
Results of hypomethylation analyses. Identification of DMRs and KEGG pathway analysis. (A) Relative DNA methylation levels of miPS-LLCcm cells, Ptdc cells and LMN cells compared to miPSCs. miPSCs vs. miPS-LLCcm cells (left), miPSCs vs. Ptdc cells (middle) and miPSCs vs. LMN cells (right). (B-D) Numbers of DMRs hypo- and hyper-methylated in each chromosome are depicted and KEGG pathways nominated with the number of hypomethylated genes. (B) miPSCs vs. miPS-LLCcm cells, (C) miPSCs vs. Ptdc cells and (D) miPSCs vs. LMN cells.