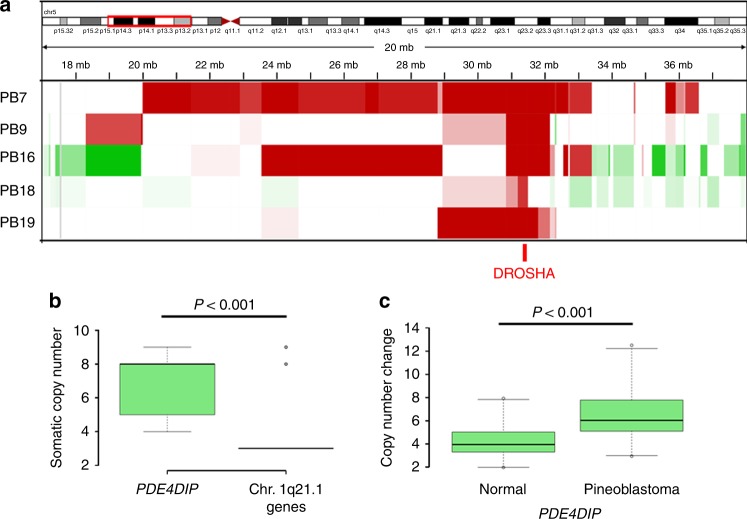
Fig. 3.
Focal copy-number aberrations in pineoblastoma. a Homozygous deletions of DROSHA were identified in five pediatric pineoblastomas. For four out of five cases with DROSHA deletion, the deletion is about 1.300–3.755 kb including the whole DROSHA gene. For one case, the deletion seems to be only 300 kb, spanning exons 11–35 of DROSHA. b, c We identified microduplication of PDE4DIP in seven pineoblastomas. b Whole-genome sequencing (n = 2) shows higher than expected copy number difference between PDE4DIP and neighboring genes in 1q21.1 region in pineoblastomas (t-test p value < 0.001). c Copy number of PDE4DIP in pineoblastoma is significantly increased compared to the number of PDE4DIP copies in normal DNA by ddPCR (average number of PDE4DIP copies in pineoblastoma six, vs. four in normal controls, paired t-test between normal and tumors <0.001). Genes neighboring PDE4DIP were not amplified by ddPCR confirming that the microduplication was limited to PDE4DIP only