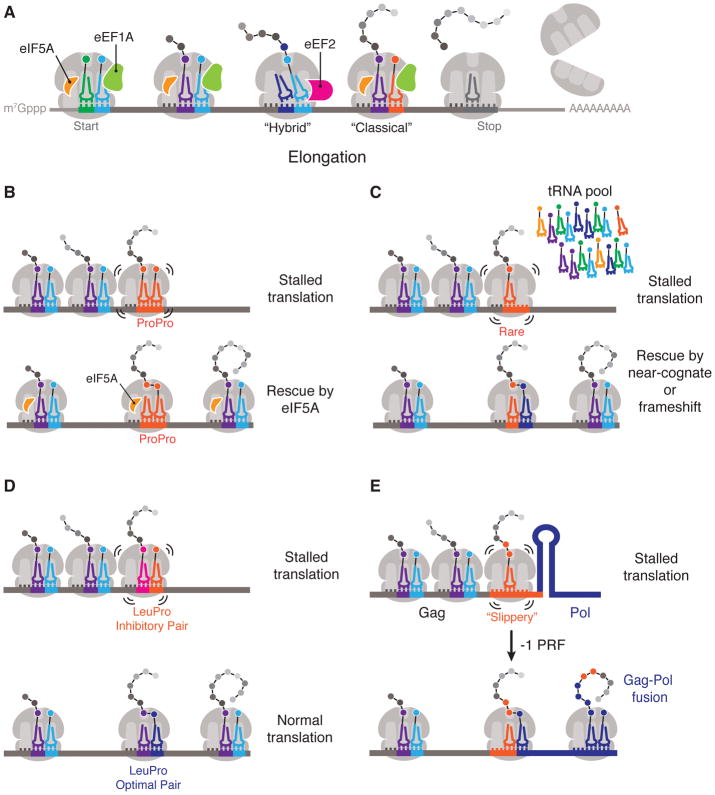
Figure 2. Translation elongation and resolution of ribosome stalling.
(A) Overview of translation elongation. Aminoacyl-tRNAs are delivered to the ribosome in complex with elongation factor 1-alpha (eEF1A) and GTP (not shown). Peptide bond formation occurs and the tRNAs are positioned in a ‘hybrid’ state with respect to the ribosome subunits. Subsequent translocation driven by elongation factor 2 (eEF2) causes tRNA repositioning from a hybrid state to classical state, creating an open A site for the next incoming aminoacyl-tRNA. eIF5A, which is a small protein that binds in the ribosomal E site, stimulates catalysis in the peptidyl transferase center throughout translation elongation. (B) Ribosome stalling due to slow peptidyl-transfer kinetics (such as during the formation of Pro–Pro) is rescued by eIF5A, which promotes peptide-bond formation. (C) Ribosome stalling caused by poor A-site occupancy resulting from poorly-expressed tRNAs (represented in figure) or poor tRNA aminoacylation can be rescued by mis-incorporation of near-cognate tRNAs or by frameshifting (represented as conversion of orange to purple). (D) Ribosome stalling can be caused by certain consecutive tRNA–codon pair orders that are sub-optimal (pink and orange) relative to synonymous pairs (purple and blue). (E) Ribosome stalling caused by mRNA secondary structures can be resolved by programmed ribosomal frameshifting (PRF) at adjacent slippery sequences. The example illustrates the -1 PRF that is required for translation of the Gag-Pol fusion protein of human immunodeficiency virus.