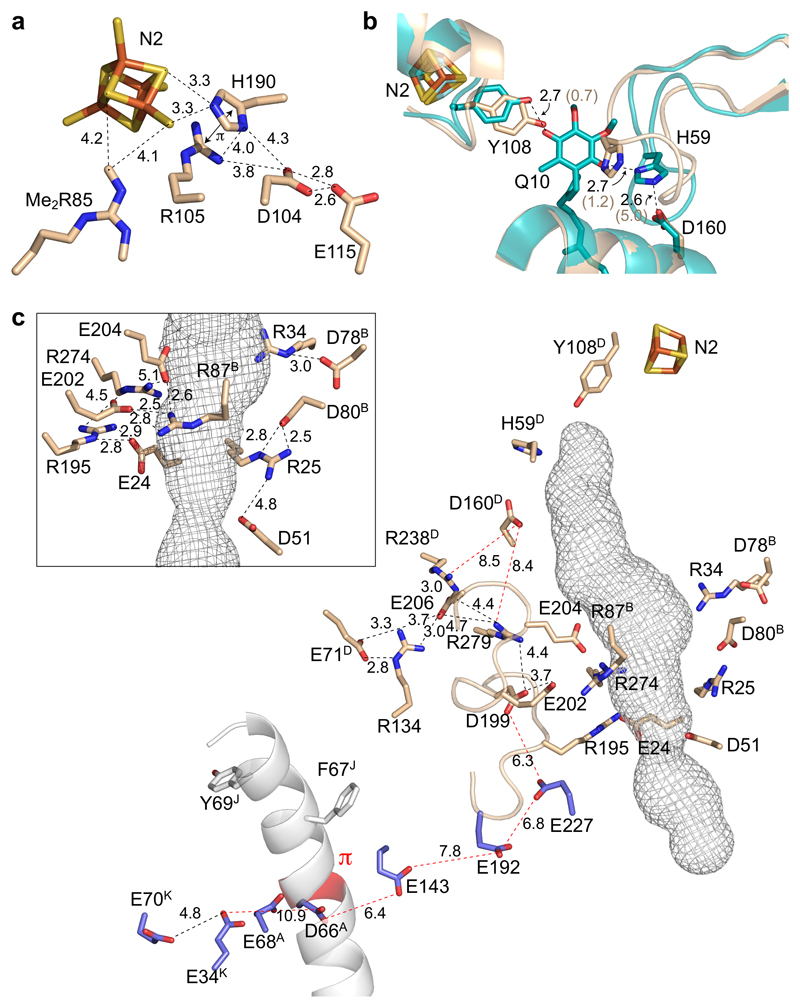
Figure 6. The environment of cluster N2, the ubiquinone-binding channel and the E-channel in active mouse complex I.
a) Cluster N2 and nearby key residues in NDUFS2. Dimethyl-Arg85 and Arg105 are positively charged, His190 is the redox-Bohr group that changes protonation state upon N2 reduction41,42, and Asp104 and Glu115 may transfer protons to/from His190. b) The structure around the ubiquinone-headgroup binding site with key residues in NDUFS2. The structure of active mouse complex I (wheat) is overlaid on the modeled structure of bovine complex I with ubiquinone-10 bound (teal)45 in which hydrogen bonds are present between the headgroup and Tyr108 and His59, and His59 and Asp160. Distances in brackets are for the active mouse structure. c) The ubiquinone-binding channel (mesh) in active mouse complex I was predicted using CAVER 3.054 with a 1.4 Å probe radius. The key residues from panel b are shown with charged residues in the vicinity of the site (inset, charged residues around the center of the channel and their interactions). The main figure highlights the carboxylate-rich ND1 TMH 5-6 loop and the E channel residues (blue) that connect the ubiquinone-binding site to the π-helix present in ND6 in the deactive enzyme, and to charged residues in the central membrane plane7. Long distances of interest are in red, and all residues are in ND1 unless otherwise indicated (B - NDUFS7, D - NDUFS2, J - ND6 and K - ND4L). All distances are in Å.