Abstract
Current blood biomarkers are suboptimal in detecting drug-induced liver injury (DILI) and predicting its outcome. We sought to characterize the natural variabilty and performance characteristics of fourteen promising DILI biomarker candidates. Serum or plasma from multiple cohorts of healthy volunteers (n=192 and =81), subjects who safely took potentially hepatotoxic drugs without adverse effects (n=55 and =92) and DILI patients (n=98, =28, and =143) were assayed for microRNA-122 (miR-122), glutamate dehydrogenase (GLDH), total keratin 18 (K18), caspase cleaved K18 (ccK18), glutathione S-transferase alpha (GSTα), alpha fetoprotein (AFP), arginase-1 (ARG1), osteopontin (OPN), sorbitol dehydrogenase (SDH), fatty acid binding protein (FABP1), cadherin-5 (CDH5), macrophage colony stimulating factor receptor (MCSFR), paraoxonase 1 (PON1, normalized to prothrombin protein), and leucocyte cell-derived chemotaxin-2 (LECT2). Most candidate biomarkers were significantly altered in DILI cases compared to healthy volunteers. GLDH correlated more closely with gold standard alanine aminotransferase (ALT) than miR-122 and there was a surprisingly wide inter- and intra-individual variability of miR-122 levels among the healthy volunteers. Serum K18, OPN, and MCSFR levels were most strongly associated with liver-related death or transplant within 6 months of DILI-onset. Prediction of prognosis among DILI patients using Model for End-stage Liver Disease (MELD) was improved by incorporation of K18 and MCSFR levels. Conclusion: GLDH appears to be more useful than miR-122 in identifying DILI patients. K18, OPN and MCSFR are promising candidates for prediction of prognosis during an acute DILI event. Serial assessment of these biomarkers in large prospective studies will help further delineate their role in DILI diagnosis and management.
Keywords: Predictive Safety Testing Consortium (PSTC), Safer and Faster Evidence-based Translation (SAFE-T), Drug-Induced Liver Injury Network (DILIN), hepatotoxicity, reference interval
Drug-induced liver injury (DILI) is a serious concern for patients, clinicians and pharmaceutical companies, accounting for over half of the acute liver failure cases observed in Western countries [1, 2]. Current detection and assessment of DILI relies on measurement of analytes that have been utilized for decades. Serum enzyme activities of aminotransferases, [alanine aminotransferase (ALT) and aspartate aminotransferase (AST)] and alkaline phosphatase (ALP) are quantified as measures of hepatocellular or cholestatic injury, respectively, whilst serum total bilirubin (TBIL) concentration is frequently used to assess global liver function. However, alterations in these biomarkers are not mechanistically informative, can occur for a variety of reasons unrelated to hepatic injury [3–5], and can be observed with drugs that do not have the potential to cause clinically significant DILI [6, 7]. They also are not specific or selective for DILI versus other causes of liver injury. Moreover, it is not currently possible to distinguish benign liver chemistry elevations from those that could lead to liver failure. Hence, even mild aminotransferase elevations can increase liver safety concern, especially in early clinical trials. A combination of an injury marker (aminotransferases) and a functional marker (TBIL) in the absence of cholestasis and when other causes have been excluded (i.e., Hy’s Law case criteria) is a widely accepted prognostic model for DILI outcome [8]. However, most of the patients that meet Hy’s Law case criteria will not require a liver transplant or die. Ongoing efforts are exploring methods to improve severity prediction utilizing traditional tests [9]. Nevertheless, there is a clear unmet need for biomarkers that are mechanistically informative and sensitive, as well as specific for prediction of DILI progression or resolution.
Substantial resources have been committed to understanding DILI and advancing candidate biomarkers that add value to traditional liver tests. Particularly, the Critical Path Institute’s Predictive Safety Testing Consortium (PSTC) in the United States and the Safer and Faster Evidence based Translation (SAFE-T) consortium within the Innovative Medicines Initiative (IMI) in Europe were leading major efforts to validate and qualify novel DILI biomarkers [10]. The Drug Induced Liver Injury Network (DILIN) is a multi-center network in the United States created to prospectively biobank blood and tissue specimens from patients who have experienced DILI [11]. Each subject in this registry has undergone an unprecedented degree of phenotyping and most have at least six-month follow-up data aiding assessments of long-term outcomes and prognosis. Given the overlapping goals of these three organizations, a cross functional collaboration was established to study performance characteristics of candidate DILI biomarkers.
Candidate DILI biomarkers have been identified in preliminary evaluations. However, clinical application of these candidate markers requires robust performance for DILI detection and prognosis compared to current standards. Further, it is critical that normal reference intervals be established for these biomarkers against which data from patients can be measured. Herein the results from an international collaborative effort among PSTC, SAFE-T, and DILIN are presented. Alpha fetoprotein (AFP), arginase 1 (ARG1), cadherin 5 (CDH5), fatty acid binding protein 1 (FABP1), glutathione S transferase alpha (GSTα), total keratin 18 (K18), caspase cleaved (cc)K18, macrophage colony stimulating factor receptor (MCSFR), osteopontin (OPN), glutamate dehydrogenase (GLDH), leucocyte cell-derived chemotaxin-2 (LECT2), paraoxonase 1 (PON1, normalized to prothrombin protein), sorbitol dehydrogenase (SDH), and microRNA-122 (miR-122) were assayed in serum or plasma from two cohorts of normal healthy subjects, two cohorts of patients that safely took potentially hepatotoxic drugs, and three cohorts of DILI patients (Table 1). DILI cohorts included patients who at 6 months had recovered completely, had persistent DILI or who died or required a liver transplant due to the DILI event within 6 months of DILI-onset.
Table 1.
Candidate Biomarkers
| Candidate Biomarker | Physiological Function | Tissue Localization | Potential Utility in DILI | Cohorts Analyzed |
|---|---|---|---|---|
| AFP | plasma protein thought to be the fetal form of albumin | high levels in liver progenitor cells | regeneration (progenitor cells) | DILIN, PSTC, SAFE-T |
| ARG1 | hydrolase enzyme that catalyzes the hydrolysis of arginine to urea and orthinine | high levels in liver; lower levels in erythrocytes, kidney and brain | cell injury/death | DILIN, PSTC, SAFE-T |
| CDH5 | calcium-dependent transmembrane adherens junction protein important for endothelial cell integrity and cell-cell adhesion | broad localization including liver | susceptibility | DILIN, PSTC, SAFE-T |
| FABP1 | protein involved in binding and transport of fatty acid | high levels in the liver; lower levels in kidney and gastrointestinal tract | cell injury/death | DILIN, PSTC, SAFE-T |
| GSTα | phase II detoxification enzyme that catalyzes the conjugation of glutathione with various electrophiles | high levels in the liver and multiple tissues | cell injury/death | DILIN, PSTC, SAFE-T |
| K18/ccK18 | type I intermediate protein expressed in epithelial cells responsible for cell structure and integrity. Caspase cleavage results in a fragmented form of protein (ccK18). | broad localization including liver | cell injury/death, mechanism | DILIN, PSTC, SAFE-T |
| MCSFR | receptor on macrophages/monocytes for CSF, a cytokine that controls the proliferation, differentiation, and function of macrophages | broad localization including liver | inflammation | DILIN, PSTC, SAFE-T |
| OPN | phosphoprotein involved in migration/infiltration of inflammatory and cancer cells | broad localization including liver | inflammation, regeneration (progenitor cells) | DILIN, PSTC, SAFE-T |
| GLDH | mitochondrial matrix protein that catalyzes the conversion of 2-oxoglutarate to L-glutamate | high levels in the liver; lower levels in kidney and brain | cell death, mechanism | PSTC, SAFE-T |
| LECT2 | protein involved in the recruitment of neutrophils | high expression in the liver; lower expression in the testis | regeneration (hepatocytes) | PSTC, SAFE-T |
| PON1 | HDL-associated enzyme that participates in paraoxonase, arylesterase, and dyazoxonase activites. Useful in diagnosis of NAFLD and NASH when normalized to prothrombin. | produced in liver, released constitutively into circulation | function | PSTC, SAFE-T |
| SDH | enzyme involved in carbohydrate metabolism that converts sorbitol into fructose | high levels in the liver, kidney, and testis; lower levels in multiple tissues | cell injury/death | PSTC, SAFE-T |
| miR-122 | liver-specific miRNA that post-transcriptionally regulates mRNA involved in processes including hepatocyte differentiation and lipid/cholesterol metabolism | high levels in the liver | cell injury/death | PSTC, SAFE-T |
Abbreviations: AFP, alpha fetoprotein; ARG1, arginase 1; CDH5, cadherin 5; FABP1, fatty acid binding protein 1; GST-α, glutathione S transferase alpha; K18, total keratin 18; ccK18, caspase cleaved K18; MCSFR, macrophage colony stimulating factor receptor; OPN, osteopontin; GLDH, glutamate dehydrogenase; LECT2 leukocyte cell derived chemotaxin 2; PON1, paroxonase 1 normalized to prothrombin protein; SDH, sorbitol dehydrogenase; miR-122, microRNA-122; HDL, high-density lipoprotein; NAFLD, non-alcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; DILIN, Drug-Induced Liver Injury Network; PSTC, Predictive Safety Testing Consortium; SAFE-T, Safer and Faster Evidence-based Translation
MATERIALS AND METHODS
Human Subject Sample Collection
Demographic data for the respective population cohorts can be found in Supplemental Tables 1–3. All studies were conducted in accordance with protocols approved by local Institutional Review Boards and written consent was received from all participants. Inclusion and exclusion criteria for each cohort is described in more detail in Supplemental Methods. The primary causative drug implicated for all DILI patients is listed in Supplemental Table 4. Following isolation of serum or plasma, samples were biobanked at −80°C until analysis and may have been frozen for multiple years.
Biomarker Analysis
The 14 candidate biomarkers that were quantified in this study are listed in Table 1. Detailed methods for biomarker quantification can be found in Supplemental Methods. Briefly, serum or plasma traditional biomarker levels were quantified by clinical chemistry at the local institutions. Serum or plasma candidate biomarker levels were quantified in assays designed by or optimized by Natural and Medical Sciences Institute (NMI, Reutlingen, Germany) or at contract laboratories. Detailed information regarding validation parameters for all assays used in this study can be found in Supplemental Table 5.
Statistical Analysis
Descriptive statistics, median with interquartile range (IQR), were used to describe continuous variables, and frequency and percent were used to describe categorical variables. All statistical analyses were performed using JMP Genomics v8.1 or SAS software (SAS, Cary, NC) or GraphPad Prism 7.01 (La Jolla, CA). Biomarker distribution was visualized and the majority of displayed a log normal distribution. For consistency, the absolute value of all biomarkers, with the exception of the Apoptotic Index (AI), were log transformed for statistical analyses. Statistical significance was considered p<0.05.
Reference Interval Determination
PSTC healthy volunteer data and SAFE-T Tel Aviv healthy volunteer data were analyzed for the determination of reference intervals. The reference interval lower limit of normal (LLN) and ULN was defined by the 5th and 95th percentile of the population, respectively, using a mixed model approach and fitted a random subject term. For two of the markers (ccK18 and GSTα) a substantial number of values were below the lower limit of quantification (LLoQ), so a maximum likelihood estimate for a truncated log-normal distribution was used to estimate the ULN. Because ~90% of the data for K18 was below the LLoQ, only the ULN was calculated by a nonparametric method. PSTC collected three serial biomarker measurements in all subjects and a mixed model was used to obtain the variance components for inter-and intra-individual variation assuming log-normal distribution. The reference interval was obtained using the estimated mean and standard deviation for the log-normal distribution.
Biomarkers of DILI Detection
Receiver operator characteristic (ROC) curve analysis was utilized to determine the performance of traditional and candidate biomarkers for detection of DILI patients. Biomarkers were considered predictive of DILI if both the ROC AUC and the lower end of the 95% confidence interval (CI) >0.5. All healthy volunteer and patient datasets (PSTC, SAFE-T, and DILIN) were used for this analysis. The relationship of liver-specific GLDH and miR-122 to ALT was examined. Correlation of GLDH and miR-122 with ALT was determined using Pearson’s r.
Biomarkers of DILI Prognosis
Accurate outcome assessments were available only for the DILIN subjects. ROC curve analysis was utilized to determine which biomarkers measured in the initial DILIN sera could significantly predict which patients died/required a liver transplant or developed unresolved DILI. For a detailed description of this analysis, refer to Supplemental Methods.
RESULTS
Biomarker Levels in Healthy Volunteers
The natural variation of candidate biomakers was explored in two cohorts of healthy volunteers (see Supplemental Tables 1 and 2). Multiple samples returned values below the LLoQ for AFP, ccK18, GSTα, K18, SDH, and miR-122. The geometric mean, inter-subject variation and reference interval (5th and 95th percentiles) for each biomarker are presented in Table 2. Due to the large number of samples with K18 values that fell below the LLoQ, only the estimated 95th percentile was calculated for this biomarker. In both healthy volunteer cohorts, inter-subject variability of miR-122 was also high [% coefficient of variation (CV) of 90.89 and 213.51 in the PSTC and SAFE-T cohorts, respectively]. Further, miR-122 also showed substantial intra-subject variability in the PSTC cohort (intrasubject %CV of 93.56) and this appeared to be most prominent among black individuals (Figure 1).
Table 2.
Biomarker Reference Intervals in Healthy Volunteers
| Biomarker | Unit | Matrix | PSTC (n=81)* | SAFE-T (n=192) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Est. Geometric Mean | LLN | ULN | Inter-Subj %CV | Intra-Subj %CV | Est. Geometric Mean | LLN | ULN | Inter-Subj %CV | |||
| Est. 5th Percentile | Est. 95th Percentile | Est. 5th Percentile | Est. 95th Percentile | ||||||||
| AFP | ng/ml | serum | 0.68 | 0.24 | 1.98 | 61.53 | 31.93 | 0.99 | 0.28 | 3.54 | 90.21 |
| ARG1 | ng/ml | serum | 7.63 | 3.00 | 19.46 | 46.03 | 37.46 | 35.97 | 18.38 | 70.38 | 42.57 |
| ccK18 | U/L | serum | 90.65 | 31.59 | 260.16 | 70.97 | 34.76 | 139.99 | 52.46 | 373.55 | 65.39 |
| CDH5 | ng/ml | serum | 2798.89 | 1853.77 | 4225.87 | 18.00 | 17.69 | 2287.79 | 1222.52 | 4281.33 | 39.52 |
| FABP1 | ng/ml | serum | 6.91 | 3.29 | 14.55 | 32.75 | 32.86 | 9.21 | 4.57 | 18.54 | 44.55 |
| GLDH | U/L | serum | 2.71 | 1.01 | 7.24 | 52.74 | 34.53 | 3.00 | 0.95 | 9.51 | 79.68 |
| GSTα | ng/ml | serum | 6.31 | 0.68 | 60.00 | 119.54 | 71.86 | 6.61 | 0.71 | 64.11 | 172.57 |
| LECT2 | ng/ml | plasma | 252.27 | 142.07 | 447.96 | 28.64 | 20.97 | 177.96 | 84.74 | 373.74 | 47.50 |
| miR-122 | copies/μl | serum | 2152.98 | 347.05 | 13356.52 | 90.89 | 93.56 | 3173.64 | 368.02 | 27367.61 | 213.51 |
| MCSFR | ng/ml | plasma | 334.81 | 196.1 | 571.64 | 30.08 | 13.89 | 306.67 | 175.98 | 534.39 | 34.75 |
| OPN | ng/ml | serum | 4.13 | 1.66 | 10.31 | 52.15 | 26.61 | 6.54 | 2.68 | 15.99 | 58.56 |
| PON1 | ng/μg | plasma | 4.91 | 2.02 | 11.93 | 44.16 | 34.57 | 9.44 | 4.18 | 21.35 | 52.81 |
| SDH | U/L | serum | 3.02 | 1.18 | 7.75 | 43.43 | 41.01 | 1.79 | 0.79 | 7.17 | 101.57 |
| K18** | U/L | serum | 121.35 | 151.14 | |||||||
Three serial collections were collected for each individual. The mean value for each individual was used for all statistical analyses with the exception of intra-individual %CV
90% of K18 data was below the lower limit of quantification, therefore only an upper reference interval was determined
Abbreviations: PSTC, Predictive Safety Testing Consortium; SAFE-T, Safer and Faster Evidence-based Translation; Est, established; LLN, lower limit of normal; ULN, upper limit of normal; CV, coefficient of variation; CI, confidence interval AFP, alpha fetoprotein; ARG1, arginase 1; ccK18, caspase cleaved K18; CDH5, cadherin 5; FABP1, fatty acid binding protein 1; GLDH, glutamate dehydrogenase; GSTα, glutathione S transferase alpha; LECT2, leukocyte cell derived chemotaxin 2; miR-122, microRNA-122; MCSFR, macrophage colony stimulating factor receptor; OPN, osteopontin; PON1, paroxonase 1 normalized to prothrombin protein; SDH, sorbitol dehydrogenase; K18, keratin 18
Figure 1. Intra-Individual Variabilty in PSTC Cohort Observed in microR-122 (miR-122) Quantifications.

Three fasting blood samples were collected from volunteers over the course of 21 days. Greater intra-subject variability was observed in miR-122 levels amongst black subjects in this study compared to white subjects. Each bar represents an individual subject while circles represent data for miR-122 measurements 1 (white), 2 (black), and 3 (gray).
The biomarker reference intervals between PSTC and SAFE-T showed substantial overlap, although the geometric mean tended to be slightly higher in the SAFE-T cohort. ARG1 levels, however, were considerably increased in the SAFE-T cohort, compared to the PSTC cohort.
Biomarker Performance for Detecting DILI
All candidate DILI biomarkers significantly identified patients with DILI with the exception of LECT2 (Table 3). The lower CI limit of LECT2 was only 0.45, indicating that the data cannot rule out random agreement between predictions and outcome. K18, ccK18, FABP1, and GLDH had AUCs > 0.9, indicating that these biomarkers are the most accurate candidate biomarkers for the detection of DILI.
Table 3.
Biomarker Performance in DILI Identification*
| Category | Biomarker | AUC | 95% CI |
|---|---|---|---|
| Traditional | ALT | 0.990 | 0.984 – 0.996 |
| Traditional | AST | 0.975 | 0.963 – 0.987 |
| Traditional | ALP | 0.902 | 0.873 –0.930 |
| Traditional | TBIL | 0.857 | 0.821 – 0.892 |
|
| |||
| Candidate | K18 | 0.947 | 0.928 – 0.966 |
| Candidate | FABP1 | 0.916 | 0.890 – 0.941 |
| Candidate | ccK18 | 0.911 | 0.887 – 0.935 |
| Candidate | GLDH | 0.907 | 0.870 – 0.945 |
| Candidate | MCSFR** | 0.854 | 0.822 – 0.887 |
| Candidate | miR-122 | 0.831 | 0.779 – 0.883 |
| Candidate | AFP | 0.826 | 0.793 – 0.859 |
| Candidate | GSTα | 0.827 | 0.792 – 0.862 |
| Candidate | SDH | 0.819 | 0.763– 0.876 |
| Candidate | OPN | 0.758 | 0.718– 0.799 |
| Candidate | CDH5 | 0.658 | 0.614 – 0.701 |
| Candidate | PON1 | 0.612 | 0.542 – 0.682 |
| Candidate | ARG1 | 0.564 | 0.519 – 0.609 |
| Candidate | LECT2 | 0.519 | 0.450 – 0.588 |
Abbreviations: AUC, area under the curve; CI, confidence interval; ALT, alanine aminotransferase; AST, aspartate aminotransferase; ALP, alkaline phosphatase; TBIL, total bilirubin; K18, keratin 18; FABP1, fatty acid binding protein 1; GLDH, glutamate dehydrogenase; ccK18, caspase cleaved K18; MCSFR, macrophage colony stimulating factor receptor; SDH, sorbitol dehydrogenase; miR-122, microRNA-122; AFP, alpha fetoprotein; GST-α, glutathione S transferase alpha; OPN, osteopontin; PON1, paroxonase 1; CDH5, cadherin 5; ARG1, arginase 1; LECT2, leukocyte cell derived chemotaxin 2; PSTC, Predictive Safety Testing Consortium; SAFE-T, Safer and Faster Evidence-based Translation; DILI, Drug-Induced Liver Injury; DILIN, DILI Network.
Statistical data for all biomarkers was calculated using patient data from PSTC healthy volunteers (n=81), SAFE-T healthy volunteers (n=192), SAFE-T subjects that safely received DILI-eliciting compounds (n=147), SAFE-T DILI patients (n=126). DILIN patient data (n=143) was also used for all biomarkers with the exception of GLDH, miR-122, SDH, PON1, and LECT2.
DILIN measurements collected in serum. All other measurements collected in plasma.
GLDH levels showed a very strong correlation with ALT levels (r=0.88, p<0.0001; Figure 2A). miR-122 levels were also significantly correlated with levels of ALT, although the strength of the correlation was reduced compared to that for GLDH (r=0.66; p<0.0001; Figure 2B).
Figure 2. Correlation Between Levels of Alanine Aminotransferase (ALT) and Liver-Specific Biomarkers.

Correlation of ALT with glutamate dehydrogenase (GLDH; A) and microRNA-122 (miR-122; B). Data points are individual PSTC (Predictive Safety Testing Consortium) and SAFE-T (Safer and Faster Evidence-based Translation) subject samples and represent individuals that did not have drug-induced liver injury (DILI; gray) and individuals that did have DILI (black). Values are log normalized. Pearson r is shown.
Biomarker Alterations by Drug Class
The SAFE-T DILI cohorts contain data from patients with acetaminophen (APAP)-related liver injury, as well as from patients who experienced idiosyncratic DILI related to various compounds (refer to Supplemental Table 4). To determine if one or more DILI compounds/classes produces signature biomarker changes that are unique compared to APAP-related hepatotoxicity, SAFE-T DILI patient data were divided into broad drug classes (see Supplemental Methods). In general, biomarkers, including ALT, tended to be the most altered from other drug classes in APAP-related hepatotoxicity, emphasizing the acute and severe injury this compound causes (Supplemental Figure 1A). However, several biomarkers were the most elevated in flupirtine DILI. Specifically, TBIL (p<0.05 compared to APAP, antibiotics, chemo, NSAID, and other; Supplemental Figure 1B), CDH5 (p<0.05 compared to APAP, antibiotics, chemo, and other; Supplemental Figure 1C), and MCSFR (p<0.05 compared to APAP, antibiotics, chemo, NSAID, other; Supplemental Figure 1D) were significantly elevated in flupirtine-related liver injury. Flupirtine is an aminopyridine used as a non-opioid analgesic that is well recognized to cause serious DILI in Europe (not available in the US); all SAFE-T patients with flupirtine-related liver injury met Hy’s Law case criteria. When DILIN data were divided into drug categories, no significant differences were observed; however, DILIN data does not have patients with either APAP- or flupirtine-related hepatotoxicity (data not shown).
Differences between cohorts in amoxicillin with clavulanic acid (Augmentin) DILI were explored. We found that DILIN patients with Augmentin-induced hepatotoxicity had significantly elevated levels of ALT, ARG1, FABP1, GST-α, K18, and ccK18 (p<0.05 for all) compared to SAFE-T DILI patients (Supplemental Table 6).
Biomarker Performance as Prognostic Markers
Death/Transplant
The DILIN samples utilized in the present study were limited to those collected within 2 weeks of DILI onset and, per protocol, patients were tracked for at least 6 months. A small subsets of DILIN patients died/required a liver transplant (n=15) within 6 months and it was determined that the DILIN event was the cause [12]. ROC curve analysis demonstrated that traditional biomarkers including the international normalized ratio (INR), AST, and TBIL were predictive of death/liver tranplant (lower 95% CI limit >0.5; Table 4). Of the subset of candidate biomarkers measured in the DILIN dataset (Table 1), elevated levels of OPN, K18, MCSFR, ccK18, FABP1, and AFP significantly predicted death/transplant. INR was found to have the strongest association with death/transplant (AUC=0.920) closely followed by OPN (AUC=0.858). The predictive biomarkers had >2X and >7X fold changes over DILI patients that did not experience liver failure and healthy volunteers, respectively (Supplemental Table 7).
Table 4.
Prognostic DILI Biomarkers for Death/Liver Transplant*
| Category | Biomarker | AUC | 95% CI | Value at Youden’s J* |
|---|---|---|---|---|
| Traditional | INR | 0.920 | 0.864–0.977 | 0.47 |
| Traditional | TBIL | 0.821 | 0.733–0.909 | 5.57 |
| Traditional | AST | 0.7 | 0.587–0.814 | 5.05 |
| Traditional | ALT | 0.606 | 0.433–0.78 | 6.68 |
| Traditional | ALP | 0.597 | 0.433–0.76 | 5.01 |
|
| ||||
| Candidate | OPN | 0.858 | 0.759–0.957 | 3.38 |
| Candidate | K18 | 0.832 | 0.737–0.927 | 7.98 |
| Candidate | MCSFR | 0.775 | 0.654–0.896 | 6.94 |
| Candidate | ccK18 | 0.778 | 0.676–0.881 | 6.96 |
| Candidate | FABP1 | 0.721 | 0.608–0.833 | 4.21 |
| Candidate | AFP | 0.687 | 0.566–0.809 | 1.57 |
| Candidate | CDH5 | 0.623 | 0.498–0.748 | 8.01 |
| Candidate | ARG1 | 0.588 | 0.436–0.741 | 3.47 |
| Candidate | GST-α | 0.536 | 0.359–0.713 | 6.88 |
|
| ||||
| Candidate | AI | 0.761 | 0.627–0.895 | 0.37 |
All values with the exception of AI are log normalized. Youdin’s J is a statistic that estimates the probability of an informed decision.
Abbreviations: AUC, area under the curve; CI, confidence interval; INR, international normalized ratio; TBIL, total bilirubin; AST, aspartate aminotransferase; ALT, alanine aminotransferase; ALP, alkaline phosphatase; OPN, osteopontin; K18, cytokeratin 18; MCSFR, macrophage colony stimulating factor receptor; ccK18, caspase cleaved K18; FABP1, fatty acid binding protein 1; AFP, alpha fetoprotein; ARG1, arginase 1; CDH5, cadherin 5; GST-α, glutathione S transferase alpha; AI, apoptotic index
The values of K18 and ccK18 measured in 98 DILIN patients enabled the calculation of an Apoptotic Index (AI, see Supplemental Methods for details) and AI (ccK18:K18 ratio) was also explored as a prognostic biomarker of death/liver tranplant (Table 4). Although both K18 and ccK18 levels were elevated in patients that experienced death/liver transplant, compared to patients that did not, the AI was significantly reduced in patients who died/required a liver transplant (Figure 3A–C). An AI was additionally calculated in 64 SAFE-T patients with DILI. Patients with flupirtine-related DILI did not have a significantly different mean AI, compared to patients with APAP-induced liver injury or DILI associated with other compounds (Figure 3D).
Figure 3. Assessment of Keratin 18 (K18) Measurements.

Differences in serum K18 (A), caspase cleaved K18 (ccK18; B), and Apoptotic Index (AI; C) between Drug-Induced Liver Injury Network patients that did not die/require a liver transplant by 6 months post-DILI-onset and those that did. Differences in AI (D) between SAFE-T patients experiencing DILI associated with flupirtine utilization and patients experiencing DILI unrelated to flupirtine. Data points represent individual patients. A dotted line for AI is drawn at 0.5, representing a score that suggests an equal contribution of apoptosis and necrosis. Values for K18 and ccK18 are log normalized. Significance is **p<0.01 and ***p<0.001.
Current prognostic models for liver transplant and death were explored to identify whether incorporation of biomarkers that passed filtering criteria (OPN, K18, MCSFR, and AFP; see Supplemental Methods for details) could improve prediction. Model of End-stage Liver Disease (MELD) score ≥ 20 was highly sensitive and MELD score ≥ 30 was highly specific for prognosis of death/transplant in the DILIN population. Incorporating K18 and MCSFR levels with MELD score (when MELD values were from 20–29; Figure 4) improved the specificity of using MELD score ≥ 20 (specificity of 0.889 when incorporating K18 and MCSFR with MELD score ≥ 20 vs. 0.738 with MELD score ≥ 20 alone; Supplemental Table 8) without reducing the sensitivity of using MELD ≥ 20 alone (sensitivity of 0.933 for both). Hy’s Law showed moderate performance for prediction of death or liver transplant in this DILIN cohort (sensitivity of 0.8 and specificity of 0.634).
Figure 4. Incorporation of Candidate Biomarkers Into Model of End-Stage Liver Disease (MELD) Score Prognostic Model.
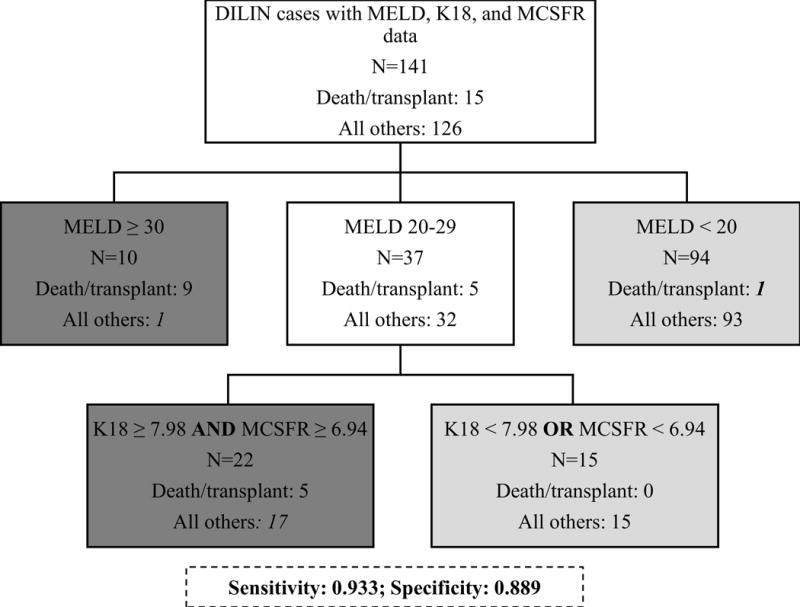
Prognostic model optimized for prediction of death/transplant in Drug-Induced Liver Injury (DILIN) patients (n=141) using the MELD score, total keratin 18 (K18), and macrophage colony stimulating factor receptor (MCSFR) measurements (sensitivity=0.933, specificity=0.889). White boxes represent branching points, light grey boxes represent patients not predicted to have an adverse outcome, and dark grey boxes represent patients predicted to die/require transplant. Numbers in italics represent false results (i.e. 18 false positives and 1 false negative).
Unresolved DILI
Nineteen patients in the DILIN cohort had unresolved DILI at their six month follow-up visit (persistently elevated ALT, AST, ALP, or TBIL over ULN with no competing etiology). Consistent with previous data, only elevated levels of ALP predicted the outcome of these patients (lower CI limit >0.5; Supplemental Table 9) [13]. Within this subset, 6 of the 19 patients with unresolved DILI had ALP levels that were elevated at their six month follow-up visit. When data was reanalyzed to determine if candidate biomarkers could predict this subset of patients, it would found that GST-α (measured within two weeks of DILI-onset) was significantly lower in patients with prolonged ALP elevation (ROC AUC = 0.760, 95% CI = 0.509–1.0).
DISCUSSION
Traditional biomarkers of DILI may not be adequately liver-specific, offer little mechanistic insight into mode of injury, and are not sufficiently prognostic for injury outcome/resolution. In the current study, the performance of 14 candidate DILI biomarkers was explored in cohorts of healthy volunteers, patients who received known DILI-eliciting compounds without developing liver injury, and in patients who experienced DILI.
Most of the biomarker reference intervals showed sizeable overlap between the SAFE-T and PSTC healthy volunteer cohorts. ARG1 levels, however, were substantially higher in the SAFE-T compared to PSTC volunteers (Table 2). This difference could not be accounted for by differences in racial diversity or age. Additionally. each biomarker was quantified at the same facility, with the same validated assays, making technical variability an unlikely explanation to account for this difference. However, biomarker stability at −80° should be explored as a possible explanation for ARG1 population differences, given that samples were stored from 3 months to 3 years. The influence of race (primarily whites vs. African Americans) on biomarker reference ranges was explored in the PSTC cohort. Although small differences were observed (such as for ARG1), there was considerable overlap between biomarker ranges and unique reference intervals were deemed to be unnecessary in this small population.
The biomarkers examined in this study were selected based on preliminary performance data generated by SAFE-T in a small pilot cohort of DILI patients and healthy volunteers (data not shown). Therefore, it is not surprising that most of the candidate biomarkers were significantly elevated in DILI. In particular, K18, FABP1, GLDH, and ccK18 had ROC AUCs >0.90, suggesting that these biomarkers in particular may be useful when screening for DILI. The sensitivity of these biomarkers for the detection of DILI or hepatotoxicity, in general, is in agreement with previously reported data [14–20]. Biomarker differences in patients with Augmentin-induced heptotoxicity were explored between the DILIN and SAFE-T cohorts and several candidate biomarkers were found to be elevated in the DILIN patients. DILIN patients also had higher mean elevations in serum ALT, therefore DILIN patients may in general have been sicker than SAFE-T patients. Indeed, while SAFE-T enrolled patients with ALT >3X ULN, DILIN inclusion criteria specifies that patients must have an ALT >5X ULN.
Although very large elevations in serum ALT are generally believed to be liver-specific, ALT elevations are also noted following strenuous exercise and in patients with muscle diseases such as muscular dystrophy [3, 21]. A biomarker with greater liver-specificity could be valuable in the clinic when the source of an ALT elevation is uncertain. GLDH and miR-122 are highly liver-specific and are not altered in response to muscle injury [22, 23]. GLDH is a large protein found within the matrix of mitochondria, enriched in the liver [24]. The inter-individual variability of GLDH ranged from ~53% in the PSTC cohort to ~80% in the SAFE-T cohort and intra-individual variability was minimal (35%). In contrast, there was significant inter-individual variability of miR-122, a liver-specific miRNA that makes up as much as 70% of hepatic miRNA content [25], in both cohorts (~91 and 213% in the PSTC and SAFE-T cohorts, respectively) and remarkable intra-individual variability, most evident among African Americans, was also observed (~94%).
MiR-122, unlike other candidate biomarkers, may not simply be leaked passively from injured cells, although this is believed to be the primary mechanism following injury. Instead, evidence suggests that miR-122 can be released actively from the liver, at least in part within extracellular vesicles, in response to stress [26]. For example, it has recently been shown that in response to stimuli and in the absence of overt hepatocyte death, miR-122 can be released and can modulate activation of innate immune cells or directly regulate kidney release of erythropoetin [27, 28]. The variability that is inherent to miR-122 levels not only between individuals but also within the same individual therefore likely results from physiologic processes unrelated to damage to the liver. The complexity of the assay (relative to other biomarker assays), and the lack of a universally accepted method for data normalization, may also contribute to the variability observed. For these reasons, the PSTC has recently deprioritized pursuit of miR-122 as a liver-specific biomarker, in favor of GLDH which is measured by routine clinical chemistry.
GLDH demonstrated enhanced correlation with ALT levels and improved performance for detection of DILI, as compared to miR-122. Previous research explored GLDH as a biomarker of hepatocellular necrosis in patients with liver impairment and found it to be superior to other candidate biomarkers (miR-122 was not evaluated in that study) [18]. Other studies have also demonstrated that GLDH is elevated in patients with APAP-related toxicity [29, 30]. Furthermore, GLDH has been proposed as a potentially early indicator of recovery from DILI due to the fast elimination of GLDH observed in subjects recovering from accidental APAP overdose with persisting high levels of ALT [14].
It should be noted that in spite of the large inter- and intra-individual variations in serum levels of miR-122 that we report here, miR-122 appears to be useful in predicting liver injury after APAP overdose. A recent study found that miR-122 demonstrated the highest performance for prediction of APAP-induced acute liver injury in a large cohort of overdose patients with normal ALT levels at presentation confirming results of an earlier study [14, 31]. In contrast, GLDH was not useful in this context. Because the method for quantitation of miR-122 in these studies differed from ours, it is unclear if the levels of miR-122 in patients susceptible to APAP injury were simply above the range of inter-and intra-individual variation meaured in the healthy volunteers in our study. While the assumption is that elevated serum levels of miR-122 reflect early hepatocyte stress or injury due to APAP, given the increasing appreciation of the physiological roles of miR-122 it is possible that individuals with high baseline serum levels of miR-122 are more susceptible to APAP injury.
The prognostic performance of GLDH and miR-122 was not determined in the current study because these biomarkers were not measured in the DILIN patient cohort (due to sample volume limitatinos) where outcome data was systematically collected. However, semi-quantitative measurements of miR-122 have previously been conducted in a subset of DILIN patients [32]. In contrast to the data observed in APAP-induced hepatotoxicity, reduced serum levels of miR-122 and albumin were observed in patients that died within six months of DILI onset, compared to patients that recovered. Collectively, these data suggest that both miR-122 and GLDH likely have utility in predicting and managing DILI and factors related to extent of injury at serum collection time and biomarker half life may influence the interpretation of biomarker alterations. Research exploring the kinetics of these biomarkers in DILI may aid in interpretation of these biomarkers in the clinic.
Several biomarkers showed promise as prognostic biomarkers for death/transplant in DILI. In particular OPN, K18, and MCSFR performed well as predictors of death/transplant. Increased levels of each of these biomarkers were observed in DILIN death/transplant patients compared to all others. OPN showed the best performance for prognosis of all candidate biomarkers in DILIN patients. OPN is associated with liver regeneration due to activation of hepatic stem cells [33]. While elevated levels of AFP, which is also released from hepatic stem cells, were prognostic for death/transplant, the performance of this biomarker was reduced compared to OPN (AUC= 0.687 vs. 0.858 for AFP and OPN, respectively).
The increase in OPN observed in this study is in contrast to recent data demonstrating that patients with acute liver failure (from various etiologies) who died/received transplant had reduced levels of plasma OPN compared to those that recovered [34]. The difference between these studies may be related to the timing of sample collection. Entry into the previous study required acute liver failure to be occurring at enrollment (INR ≥ 1.5 and encephalopathy), suggesting more advanced injury than present in the current cohort.
MCSFR, another marker of inflammation, is amongst the most promising prognostic candidate biomarkers (AUC=0.775) in the DILIN data for death/transplant. MCSFR, the receptor for CSF-1, is thought to be shed from activated macrophages during DILI [35]. Interestingly, reduced levels of CSF-1 were associated with poor outcome in patients experiencing APAP-induced liver injury and this was thought to suggest that macrophages and an innate immune response are necessary for regeneration following liver injury [36]. The cause of this discrepancy is unclear, but may be related to the type of DILI examined in the current study (idiosyncratic vs intrinsic). MCSFR levels were considerably higher in SAFE-T patients experiencing flupirtine-induced hepatotoxicity compared to the 19 cases of APAP-induced hepatotoxicity, despite ALT levels being markedly higher in APAP-induced liver injury (Figure 2). Although no SAFE-T DILI patients died or required a liver transplant, all patients that experienced flupirtine-induced DILI met Hy’s Law case criteria, suggesting that increased MCSFR may be indicative of severe idiosyncratic DILI (vs. APAP-induced liver injury).
K18 also showed value as a prognostic biomarker in DILIN patients; K18 levels were elevated in patients that died or needed a liver transplant, compared to patients that survived. We found that incorporating K18 (threshold: log normalized value of 7.98) into a model that stratified risk based on MELD score cutoffs of ≥ 20 and ≥ 30 improved specificity of using MELD ≥ 20, alone, without decreasing sensitivity. The addition of MCSFR further improved the specificity, slightly. The value of K18 for prediction models of death following hepatotoxicity is in agreement with previously published literature [15, 37].
K18 is an intermediate filament found in epithelial cells, including hepatocytes. Necrosis results in passive leakage of full length K18 into circulation while cleavage of K18 by caspases results in leakage of ccK18 into circulation following apoptotis [38]. Apoptosis is thought to be a more benign form of injury because apoptosis is not believed to result in the release of damage associated molecular pattens and subsequent activation of the innate immune system [39, 40]. Determination of the ccK18:K18 ratio, the AI, is believed to reflect the proportion of cell death that can be attributed to apoptosis. DILIN patients that died or needed a liver transplant had lower AIs than patients who recovered from DILI (i.e. consistent with necrosis as the predominate form of cell death in patients that died or needed a liver transplant). Because biopsies are not routinely conducted in the clinic, validation of the AI in humans is challenging. Nevertheless, pilot data in DILIN patients suggests that the AI may be useful in predicting the degree of apoptosis vs necrosis in liver tissue [41].
It should be noted that in the DILIN cohort two weeks was the maximum time between DILI-onset (the time at which a patient’s serum liver chemistries first qualified for DILIN entry) and research blood collection, but the time between symptom onset (when known) and research blood collection varied to a larger degree, ranging from 2 to 90 days. We were unable to detect significant correlations between biomarker levels and days between symptom onset and research blood collection (r= −0.014 to 0.222; Supplemental Table 10). For example, the interval between symptom onset and determination of serum levels of K18 and MCSFR had Pearson r’s of 0.142 and 0.163, respectively. Because variation in biomarker release and clearance kinetics may significantly impact interpretation of biomarker levels, future studies should include serial samples collected over a broad range of intervals from symptom onset.
Resulting from the data presented here, both the Food and Drug Administration and the European Medicines Agency issued Letters of Support that explicitly encourage the exploratory use of selected biomarkers in drug registration trials and further development of K18, OPN, and MCSFR as potential diagnostic or prognostic DILI biomarkers [42, 43]. Further exploration of both miR-122 and GLDH as liver-specific alternatives to ALT was also encouraged by these regulatory agencies.
In summary, the large inter- and intra-subject variation in miR-122 and the recent recognition of its regulated release from the liver without hepatocyte death may complicate its interpretation in the clinic but it is likely still valuable in certain contexts such as in the setting of APAP-induced hepatotoxicity. Alternatively, MCSFR may be elevated to a greater degree in severe idiosyncratic DILI. GLDH was a sensitive biomarker for detection of DILI and should be useful in certain clinical contexts to exclude muscle injury as a source of serum biomarkers [22]. K18, FABP1, and ccK18 were also highly sensitive for DILI detection. OPN, K18, and MCSFR show promise as biomarkers that can identify those DILI patients who will succumb to the DILI event unless transplanted. The combined use of K18, MCSFR and the MELD score improved specificity without reducing the sensitivity compared to use of a MELD score of ≥20 alone.
Based on the data reported here, follow-up initiatives should include (i) further exploration of the prognostic value of the biomarkers endorsed by regulatory agencies via broad application in clinical trials with serial sample collection (ii) correlation of the mechanism of DILI with the performance of the biomarkers (e.g. intrinsic DILI versus immune activation), and (iii) assessment of the performance of biomarkers in drug-induced versus other causes of liver injury (e.g. viral hepatitis or autoimmune hepatitis). These efforts will allow the most promising biomarkers to be validated and qualified for routine use in clinical DILI assessment.
Supplementary Material
Acknowledgments
This material is also based upon work supported by the Critical Path Institute’s (C-PATH) Predictive Safety Testing Consortium (PSTC). The authors would like to acknowledge and thank the members of PSTC for their scientific, financial, and in-kind contributions that supported these research activities, as well as the input from the FDA and EMA scientists who serve as advisors.
Financial Support:
The DILIN (https://dilin.dcri.duke.edu/) is supported by the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health (NIH) as a Cooperative Agreement (U01s) under grants: U01-DK065176 (Duke), U01-DK065201 (UNC), U01-DK065184 (Michigan), U01-DK065211 (Indiana), U01DK065193 (UConn), U01-DK065238 (UCSF/CPMC), U01-DK083023 (UTSW), U01-DK083027 (TJH/UPenn), U01-DK082992 (Mayo), U01-DK083020 (USC), U01-DK100928 (Icahn). Additional funding is provided by CTSA grants UL1 RR025761 (Indiana), UL1 RR025747 (UNC), UL1 RR024986 (UMich), and UL1 TR001420 (Wake Forest).
SAFE-T received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement n° 115003, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007–2013) and EFPIA companies” in kind contribution. Additional support was received from the NIHR Nottingham Digestive Diseases Biomedical Research Unit at the Nottingham University Hospitals NHS Trust and the University of Nottingham.
Abbreviations
- DILI
drug-induced liver injury
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- ALP
alkaline phosphatase
- TBIL
total bilirubin
- PSTC
Predictive Safety Testing Consortium
- SAFE-T
Safer and Faster Evidence-based Translation Consortium
- IMI
Innovative Medicine Initiative
- DILIN
Drug-Induced Liver Injury Network
- AFP
alpha fetoprotein
- ARG1
arginase-1
- CDH5
cadherin-5
- FABP1
fatty acid binding protein 1
- GSTα
glutathione S-transferase alpha
- K18
total cytokeratin 18
- ccK18
caspase cleaved cytokeratin 18
- MCSFR
macrophage colony stimulating factor receptor
- OPN
osteopontin
- GLDH
glutamate dehydrogenase
- LECT2
leucocyte cell-derived chemotaxin-2
- PON1
paraoxonase 1 normalized to prothrombin protein
- SDH
sorbitol dehydrogenase
- miR-122
microRNA-122
- ULN
upper limit of normal
- BMI
body mass index
- NMI
Natural and Medical Sciences Institute
- IQR
interquartile range
- AI
apoptotic index
- LLN
lower limit of normal
- LLoQ
lower limit of quantification
- ROC
receiver operator characteristic
- CI
confidence interval
- CV
coefficient of variation
- AUC
area under the curve
- APAP
acetaminophen
- NSAID
non-steroidal anti-inflammatory drug
- INR
International Normalized Ratio
- MELD
Model for End stage Liver Disease
Footnotes
COLLABORATORS
Guruprasad P. Aithal
Daniel J. Antoine
Uwe Bamberger
Thomas Berg
Patricia Chandler
Heidrun Ellinger Ziegelbauer
Neil S. French
Balasz Fülöp
Thomas Knorpp
M Isabel Lucena
Thomas O. Joos
Douglas A. Keller
Johan Lindberg
Eric Maller
Eric Massana
Jessica Mwinyi
An Ngo
Yen Ngo
Kevin B. Park
Hugo Perazzo
Hannes Planatscher
Mercedes Robles-Díaz
Camilla Stephens
Maximilian Schmeding
Nicole Schneiderhan-Marra
Eckart Schott
Joachim Tillner
Christoph Wandel
Stefan Weiler
K. Rajender Reddy
Huiman Barnhart
Jiezhun Gu
Paul H. Hayashi
Jawad Ahmad
Andrew Stolz
Victor Navarro
Jay Hoofnagle
References
- 1.Ostapowicz G, Fontana RJ, Schiodt FV, Larson A, Davern TJ, Han SH, et al. Results of a prospective study of acute liver failure at 17 tertiary care centers in the United States. Ann Intern Med. 2002;137:947–54. doi: 10.7326/0003-4819-137-12-200212170-00007. [DOI] [PubMed] [Google Scholar]
- 2.Wei G, Bergquist A, Broome U, Lindgren S, Wallerstedt S, Almer S, et al. Acute liver failure in Sweden: etiology and outcome. J Intern Med. 2007;262:393–401. doi: 10.1111/j.1365-2796.2007.01818.x. [DOI] [PubMed] [Google Scholar]
- 3.Pettersson J, Hindorf U, Persson P, Bengtsson T, Malmqvist U, Werkstrom V, et al. Muscular exercise can cause highly pathological liver function tests in healthy men. Br J Clin Pharmacol. 2008;65:253–9. doi: 10.1111/j.1365-2125.2007.03001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Purkins L, Love ER, Eve MD, Wooldridge CL, Cowan C, Smart TS, et al. The influence of diet upon liver function tests and serum lipids in healthy male volunteers resident in a Phase I unit. Br J Clin Pharmacol. 2004;57:199–208. doi: 10.1046/j.1365-2125.2003.01969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruhl CE, Everhart JE. Determinants of the association of overweight with elevated serum alanine aminotransferase activity in the United States. Gastroenterology. 2003;124:71–9. doi: 10.1053/gast.2003.50004. [DOI] [PubMed] [Google Scholar]
- 6.Aithal GP, Watkins PB, Andrade RJ, Larrey D, Molokhia M, Takikawa H, et al. Case definition and phenotype standardization in drug-induced liver injury. Clin Pharmacol Ther. 2011;89:806–15. doi: 10.1038/clpt.2011.58. [DOI] [PubMed] [Google Scholar]
- 7.Harrill AH, Roach J, Fier I, Eaddy JS, Kurtz CL, Antoine DJ, et al. The effects of heparins on the liver: application of mechanistic serum biomarkers in a randomized study in healthy volunteers. Clin Pharmacol Ther. 2012;92:214–20. doi: 10.1038/clpt.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senior JR. Alanine aminotransferase: a clinical and regulatory tool for detecting liver injury-past, present, and future. Clin Pharmacol Ther. 2012;92:332–9. doi: 10.1038/clpt.2012.108. [DOI] [PubMed] [Google Scholar]
- 9.Robles-Diaz M, Lucena MI, Kaplowitz N, Stephens C, Medina-Caliz I, Gonzalez-Jimenez A, et al. Use of Hy’s law and a new composite algorithm to predict acute liver failure in patients with drug-induced liver injury. Gastroenterology. 2014;147:109–118 e5. doi: 10.1053/j.gastro.2014.03.050. [DOI] [PubMed] [Google Scholar]
- 10.Matheis K, Laurie D, Andriamandroso C, Arber N, Badimon L, Benain X, et al. A generic operational strategy to qualify translational safety biomarkers. Drug Discov Today. 2011;16:600–8. doi: 10.1016/j.drudis.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Fontana RJ, Watkins PB, Bonkovsky HL, Chalasani N, Davern T, Serrano J, et al. Drug-Induced Liver Injury Network (DILIN) prospective study: rationale, design and conduct. Drug Saf. 2009;32:55–68. doi: 10.2165/00002018-200932010-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi PH, Rockey DC, Fontana RJ, Tillmann HL, Kaplowitz N, Barnhart HX, et al. Death and liver transplantation within 2 years of onset of drug-induced liver injury. Hepatology. 2017;66:1275–1285. doi: 10.1002/hep.29283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontana RJ, Hayashi PH, Barnhart H, Kleiner DE, Reddy KR, Chalasani N, et al. Persistent liver biochemistry abnormalities are more common in older patients and those with cholestatic drug induced liver injury. Am J Gastroenterol. 2015;110:1450–9. doi: 10.1038/ajg.2015.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antoine DJ, Dear JW, Lewis PS, Platt V, Coyle J, Masson M, et al. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology. 2013;58:777–87. doi: 10.1002/hep.26294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antoine DJ, Jenkins RE, Dear JW, Williams DP, McGill MR, Sharpe MR, et al. Molecular forms of HMGB1 and keratin-18 as mechanistic biomarkers for mode of cell death and prognosis during clinical acetaminophen hepatotoxicity. J Hepatol. 2012;56:1070–9. doi: 10.1016/j.jhep.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Dear JW, Antoine DJ, Starkey-Lewis P, Goldring CE, Park BK. Early detection of paracetamol toxicity using circulating liver microRNA and markers of cell necrosis. Br J Clin Pharmacol. 2014;77:904–5. doi: 10.1111/bcp.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thulin P, Nordahl G, Gry M, Yimer G, Aklillu E, Makonnen E, et al. Keratin-18 and microRNA-122 complement alanine aminotransferase as novel safety biomarkers for drug-induced liver injury in two human cohorts. Liver Int. 2014;34:367–78. doi: 10.1111/liv.12322. [DOI] [PubMed] [Google Scholar]
- 18.Schomaker S, Warner R, Bock J, Johnson K, Potter D, Van Winkle J, et al. Assessment of emerging biomarkers of liver injury in human subjects. Toxicol Sci. 2013;132:276–83. doi: 10.1093/toxsci/kft009. [DOI] [PubMed] [Google Scholar]
- 19.Pelsers MM, Morovat A, Alexander GJ, Hermens WT, Trull AK, Glatz JF. Liver fatty acid-binding protein as a sensitive serum marker of acute hepatocellular damage in liver transplant recipients. Clin Chem. 2002;48:2055–7. [PubMed] [Google Scholar]
- 20.Mikus M, Drobin K, Gry M, Bachmann J, Lindberg J, Yimer G, et al. Elevated levels of circulating CDH5 and FABP1 in association with human drug-induced liver injury. Liver Int. 2016 doi: 10.1111/liv.13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMillan HJ, Gregas M, Darras BT, Kang PB. Serum transaminase levels in boys with Duchenne and Becker muscular dystrophy. Pediatrics. 2011;127:e132–6. doi: 10.1542/peds.2010-0929. [DOI] [PubMed] [Google Scholar]
- 22.Flanigan KM, Voit T, Rosales XQ, Servais L, Kraus JE, Wardell C, et al. Pharmacokinetics and safety of single doses of drisapersen in non-ambulant subjects with Duchenne muscular dystrophy: results of a double-blind randomized clinical trial. Neuromuscul Disord. 2014;24:16–24. doi: 10.1016/j.nmd.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Jia Y, Zheng R, Guo Y, Wang Y, Guo H, et al. Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin Chem. 2010;56:1830–8. doi: 10.1373/clinchem.2010.147850. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt ES, Schmidt FW. Glutamate dehydrogenase: biochemical and clinical aspects of an interesting enzyme. Clin Chim Acta. 1988;173:43–55. doi: 10.1016/0009-8981(88)90356-7. [DOI] [PubMed] [Google Scholar]
- 25.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–9. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 26.Holman NS, Mosedale M, Wolf KK, LeCluyse EL, Watkins PB. Subtoxic Alterations in Hepatocyte-Derived Exosomes: An Early Step in Drug-Induced Liver Injury? Toxicol Sci. 2016;151:365–75. doi: 10.1093/toxsci/kfw047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Momen-Heravi F, Bala S, Kodys K, Szabo G. Exosomes derived from alcohol-treated hepatocytes horizontally transfer liver specific miRNA-122 and sensitize monocytes to LPS. Sci Rep. 2015;5:9991. doi: 10.1038/srep09991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivkin M, Simerzin A, Zorde-Khvalevsky E, Chai C, Yuval JB, Rosenberg N, et al. Inflammation-Induced Expression and Secretion of MicroRNA 122 Leads to Reduced Blood Levels of Kidney-Derived Erythropoietin and Anemia. Gastroenterology. 2016;151:999–1010 e3. doi: 10.1053/j.gastro.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 29.McGill MR, Sharpe MR, Williams CD, Taha M, Curry SC, Jaeschke H. The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J Clin Invest. 2012;122:1574–83. doi: 10.1172/JCI59755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGill MR, Staggs VS, Sharpe MR, Lee WM, Jaeschke H. Serum mitochondrial biomarkers and damage-associated molecular patterns are higher in acetaminophen overdose patients with poor outcome. Hepatology. 2014;60:1336–45. doi: 10.1002/hep.27265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dear JW, Clarke JI, Francis B, Allen L, Wraight J, Shen J, et al. Risk stratification after paracetamol overdose using mechanistic biomarkers: results from two prospective cohort studies. Lancet Gastroenterol Hepatol. 2017 doi: 10.1016/S2468-1253(17)30266-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russo MW, Steuerwald N, Norton HJ, Anderson WE, Foureau D, Chalasani N, et al. Profiles of miRNAs in serum in severe acute drug induced liver injury and their prognostic significance. Liver Int. 2017;37:757–764. doi: 10.1111/liv.13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arai M, Yokosuka O, Fukai K, Imazeki F, Chiba T, Sumi H, et al. Gene expression profiles in liver regeneration with oval cell induction. Biochem Biophys Res Commun. 2004;317:370–6. doi: 10.1016/j.bbrc.2004.03.057. [DOI] [PubMed] [Google Scholar]
- 34.Srungaram P, Rule JA, Yuan HJ, Reimold A, Dahl B, Sanders C, et al. Plasma osteopontin in acute liver failure. Cytokine. 2015;73:270–6. doi: 10.1016/j.cyto.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersson U, Lindberg J, Wang S, Balasubramanian R, Marcusson-Stahl M, Hannula M, et al. A systems biology approach to understanding elevated serum alanine transaminase levels in a clinical trial with ximelagatran. Biomarkers. 2009;14:572–86. doi: 10.3109/13547500903261354. [DOI] [PubMed] [Google Scholar]
- 36.Stutchfield BM, Antoine DJ, Mackinnon AC, Gow DJ, Bain CC, Hawley CA, et al. CSF1 Restores Innate Immunity After Liver Injury in Mice and Serum Levels Indicate Outcomes of Patients With Acute Liver Failure. Gastroenterology. 2015;149:1896–1909 e14. doi: 10.1053/j.gastro.2015.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Bechmann LP, Jochum C, Kocabayoglu P, Sowa JP, Kassalik M, Gieseler RK, et al. Cytokeratin 18-based modification of the MELD score improves prediction of spontaneous survival after acute liver injury. J Hepatol. 2010;53:639–47. doi: 10.1016/j.jhep.2010.04.029. [DOI] [PubMed] [Google Scholar]
- 38.Caulin C, Salvesen GS, Oshima RG. Caspase cleavage of keratin 18 and reorganization of intermediate filaments during epithelial cell apoptosis. J Cell Biol. 1997;138:1379–94. doi: 10.1083/jcb.138.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauter B, Albert ML, Francisco L, Larsson M, Somersan S, Bhardwaj N. Consequences of cell death: exposure to necrotic tumor cells, but not primary tissue cells or apoptotic cells, induces the maturation of immunostimulatory dendritic cells. J Exp Med. 2000;191:423–34. doi: 10.1084/jem.191.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Basu S, Binder RJ, Suto R, Anderson KM, Srivastava PK. Necrotic but not apoptotic cell death releases heat shock proteins, which deliver a partial maturation signal to dendritic cells and activate the NF-kappa B pathway. Int Immunol. 2000;12:1539–46. doi: 10.1093/intimm/12.11.1539. [DOI] [PubMed] [Google Scholar]
- 41.Church RJ, Kullak-Ublick GA, Kleiner DE, Bonkovsky HL, Chalasani N, Fontana RJ, et al. Candidate liver safety biomarkers provide prognostic and mechanistic insights in patients with drug-induced liver injury. Hepatology. 2017;66:24A–25A. [Google Scholar]
- 42.FDA. US Food & Drug Administration Letter of Support Initiative. 6/15/17]; Available from: https://www.fda.gov/downloads/Drugs/DevelopmentApprovalProcess/UCM517355.pdf.
- 43.EMA. EMA Letter of support for drug-induced liver injury (DILI) biomarker. 6/15/17]; Available from: http://www.ema.europa.eu/docs/en_GB/document_library/Other/2016/09/WC500213479.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


