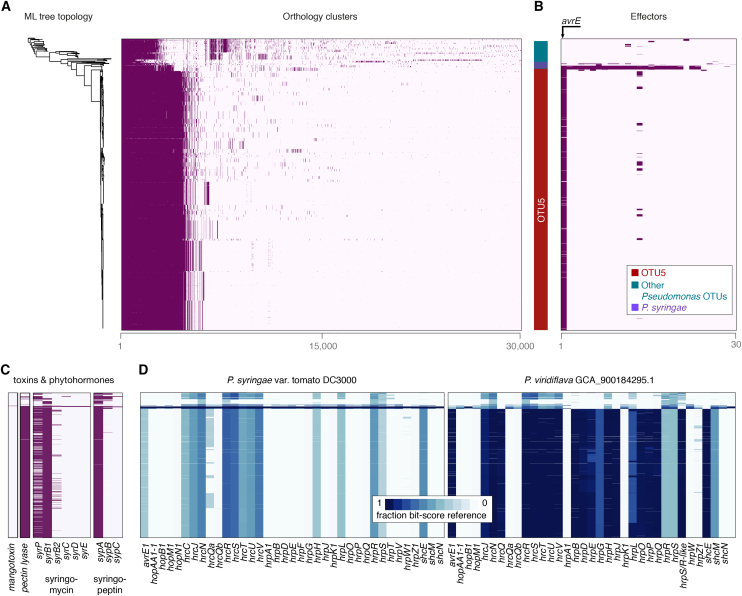
Figure 6.
OTU5 Strains Vary in Gene Content but Share an Effector
(A) ML tree topology of 1,524 isolates (for all panels) and presence (dark purple) or absence (light purple) of the 30,000 most common orthologs as inferred with panX (Ding et al., 2018). OTU5 strains share 622 ortholog groups that are found at less than 10% frequency outside OTU5.
(B) Presence/absence of 30 effector homologs. Only avrE homologs are present in more than 50 isolates.
(C) Genes for toxins and phytohormones. Only a few genomes contain the full set of genes required for synthesis of syringomycin and syringopeptin.
(D) Similarity of avrE and hrp-hrc genes to Pto DC3000 and the most similar P. viridiflava genome from NCBI. Higher values indicate greater similarity.
Related to Table S2.