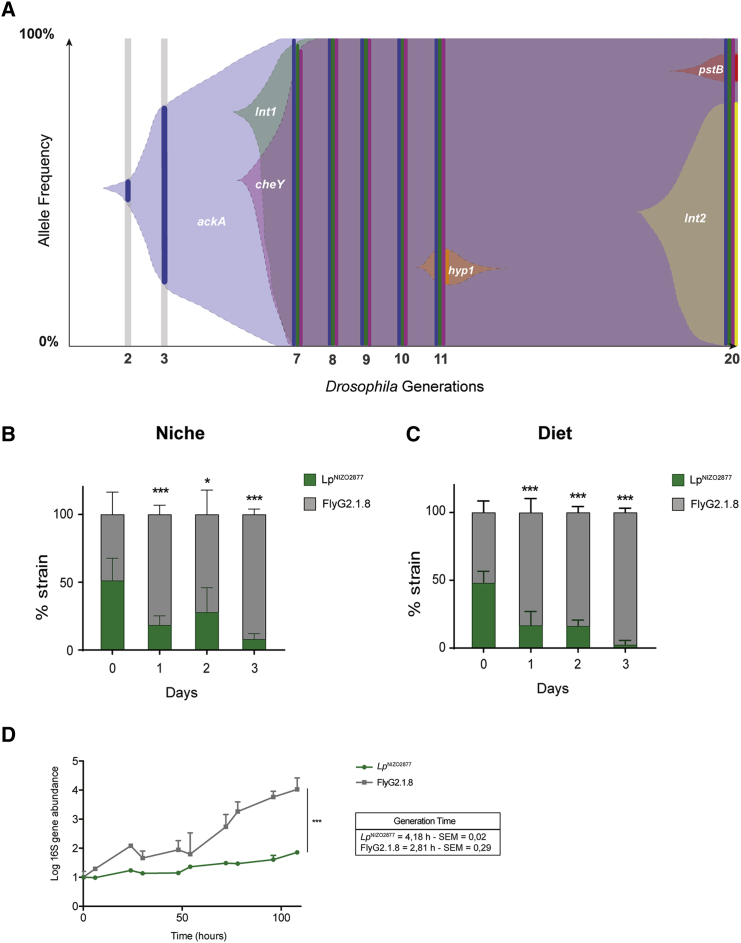
Figure 4.
LpNIZO2877-Evolved Strain Shows Higher Fitness Compared with the Ancestor
(A) Muller diagram showing the genome evolutionary dynamics of LpNIZO2877 population (replicate I) along 20 Drosophila generations. The y axis shows the percentage of the detected frequencies of each mutation (plain colors). Shaded areas represent the inferred allele frequencies. Lower axis shows the fly generation where the sampling took place.
(B and C) 1:1 competitive assay between LpNIZO2877 and LpNIZO2877-evolved strain (FlyG2.1.8) in poor-nutrient diet with Drosophila larvae (B) and without Drosophila larvae (C). Error bars represent the percentage of each strain detected in each sample (Niche or Diet) by qPCR. ∗p < 0.05, ∗∗∗p < 0.01 obtained by Student's t-test.
(D) 16S rRNA kinetics of LpNIZO2877 and FlyG2.1.8 in the Drosophila nutritional environment. Absolute quantification of the 16rRNA gene (ng/μL) has been conducted for each time point. 16S rRNA gene quantification is shown in logarithmic scale. The values have been normalized by the mean of t0 (time 0). The mean generation time (h) of each strain ± SEM is reported on the graph (see STAR Methods). The result of the non-parametric analysis of covariance (sm.ancova function in R) between the curves is reported (∗∗∗p < 0.005).