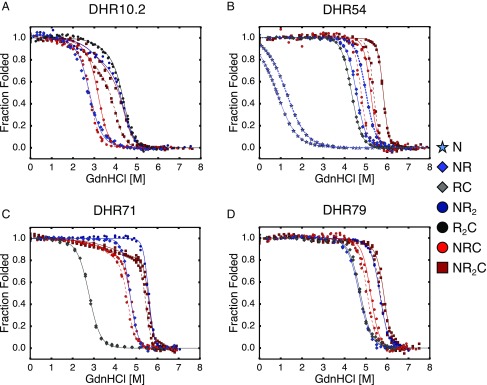
Fig. 2.
Unfolding transitions and nearest-neighbor Ising analysis of DHR proteins of different length and capping architecture. Guanidine-induced unfolding transitions for (A) DHR10.1, (B) DHR54, (C) DHR71, and (D) DHR79 were fitted with a nearest-neighbor Ising model (curves). N-capped constructs are shown in blue, C-capped constructs are shown in gray, and doubly capped constructs are shown in red. Glycerol concentrations are 0% (dash-dotted curves), 10% (solid curves), and 20% (dashed curves). For all constructs, increasing the number of repeats increases stability (based on unfolding midpoints). Conditions: 25 mM NaPO4, 150 mM NaCl, 25 °C.