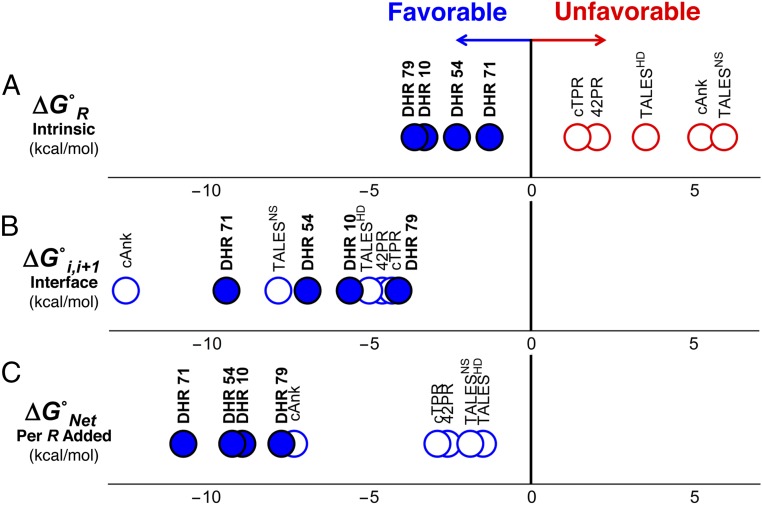
Fig. 3.
DHR repeats are intrinsically stable, unlike the repeats of naturally occurring repeat proteins. (A) Intrinsic folding and (B) interfacial coupling free energies determined by Ising analysis for DHR proteins (filled circles) and natural repeat proteins [open circles, TALESNS and TALESHD (5), 42PR (4), cANK (3), and cTPR (4)]. Unfavorable (i.e., positive) free-energy terms are in red, and favorable (i.e., negative) free energies are in blue. DHRs are stabilized by both favorable intrinsic folding and interfacial coupling free energies, whereas natural repeat proteins are destabilized by unfavorable intrinsic folding free energies, which are compensated by large favorable interfacial interactions. (C) Free energy associated with adding a single repeat to a folded array (the sum of intrinsic and interfacial free energies in A and B). Due to their favorable intrinsic folding free energies, DHR proteins are more strongly stabilized by the addition of repeats than natural repeat proteins, resulting in very high overall stability.