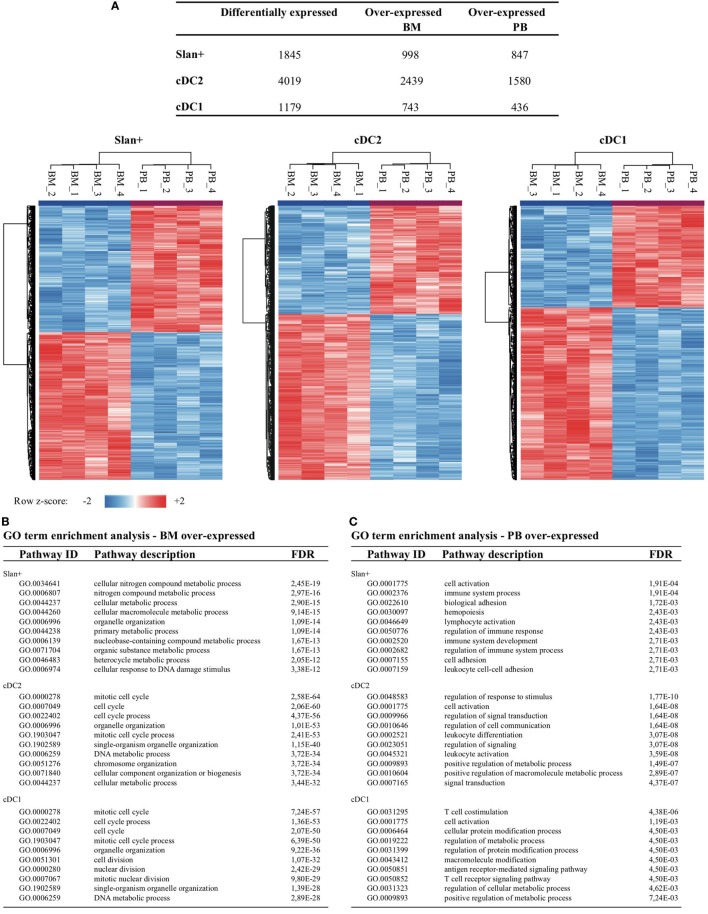
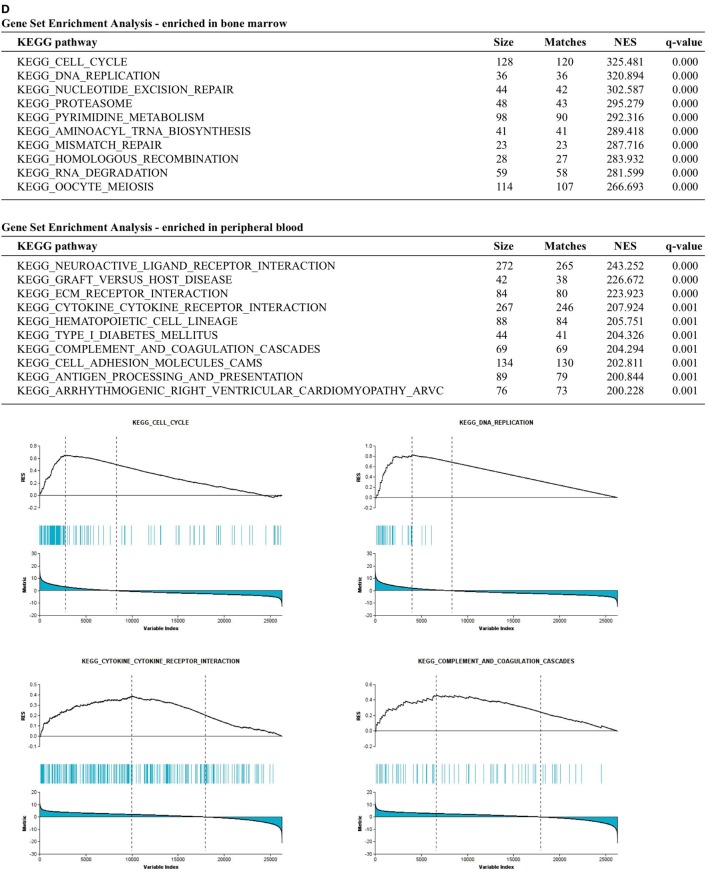
Figure 4.
Transcriptomic comparison between cell subsets of the bone marrow (BM) vs the peripheral blood (PB) compartment. Gene expression profiles were compared between the BM and PB for each individual cell subset. (A) Number of differentially expressed transcripts found between the two different compartments (e.g., slan+ in BM vs slan+ in PB). Differentially expressed genes (DEGs) were clustered per row by using z-scores for each transcript, thereby reflecting relative expression levels. Heat maps for each individual subset are shown. For each cell subset, a pathway enrichment analysis was performed in which all DEGs were used that were over-expressed in BM (B) or over-expressed in PB (C). Top 10 enriched biological processes for both compartments and each individual subset are shown. (D) Gene set enrichment analysis for BM- and PB-derived cell subsets. The top 10 of enriched KEGG pathways are shown together with four selected enrichment plots. Abbreviations: FDR, false discovery rate; GO, gene ontology; ID, identification; NES, normalized enrichment score.