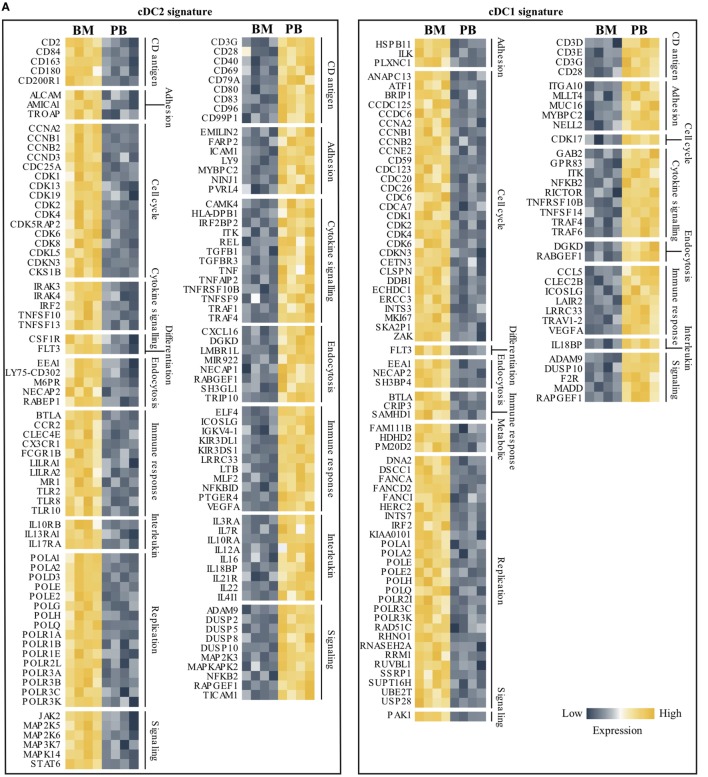
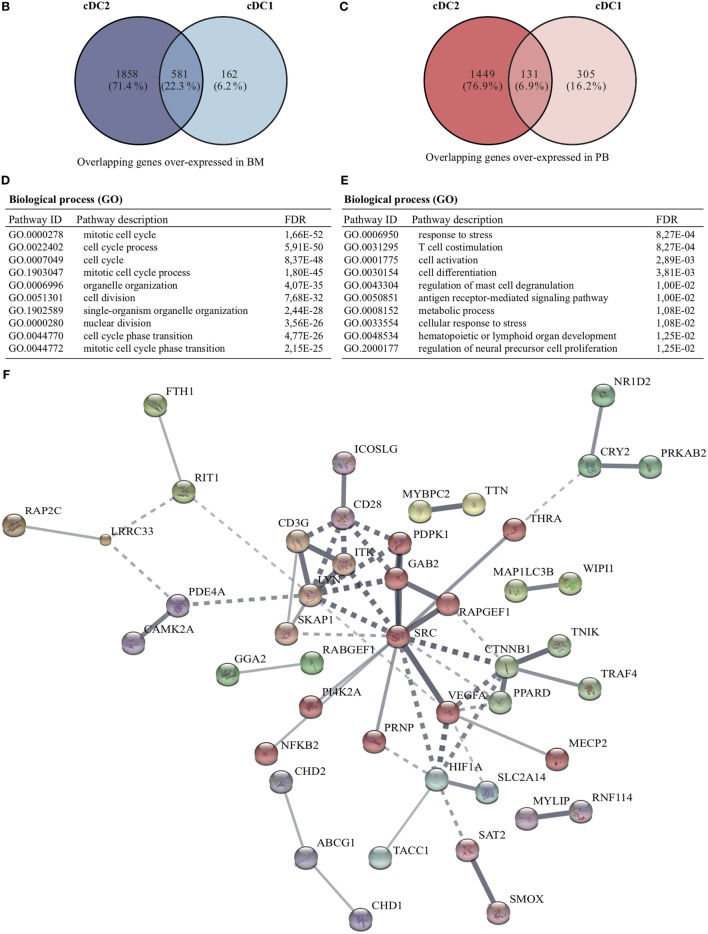
Figure 5.
Compartment-specific DC gene signatures for cDC2 and cDC1. Both myeloid DC subsets were further analyzed to find compartment-specific DC gene signatures. (A) Transcripts were filtered based on gene descriptions and GO biological process terms by using cell cycle- and immune response-related terms. Relative expression for cDC2 and cDC1 in both bone marrow (BM) as well as peripheral blood (PB) are shown for selected genes. Venn diagrams were used to obtain overlapping genes between cDC2 and cDC1 in the BM (B) and PB (C) compartment. (D) The 581 commonly over-expressed transcripts in BM-derived DCs were used in a pathway enrichment analysis. (E) Similarly, overlapping transcripts (131 in total) of PB cDC2 and cDC21 were used in the STRING pathway analysis. (F) An interaction network of the PB transcripts was generated. In total, 85 genes could be annotated. The minimum required interaction score was set to medium confidence (0.400), disconnected nodes were hidden and the thickness of the lines indicates the strength of data support. Abbreviations: FDR, false discovery rate; GO, gene ontology; ID, identification.