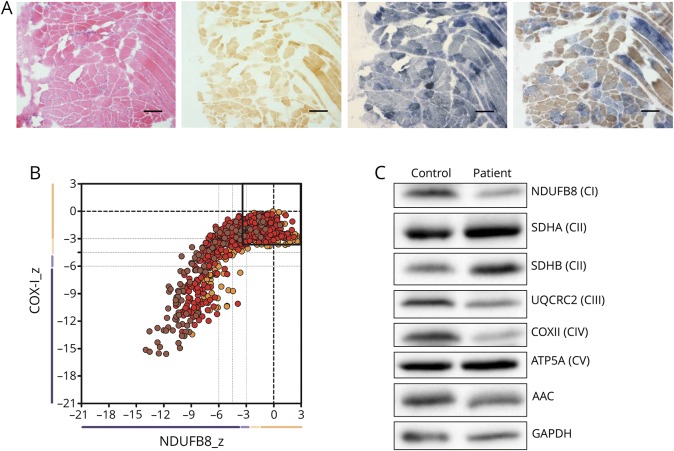
Figure 1. The de novo p.Lys33Gln mutation leads to OXPHOS defect in muscle.
(A) Histopathologic analysis of patient skeletal muscle sections showing hematoxylin and eosin (H&E) staining (far left), COX histochemistry (middle left), SDH histochemistry (middle right), and sequential COX-SDH histochemistry (far right). Scale bar = 100 µm. (B) Respiratory chain profile following quadruple oxidative phosphorylation immunofluorescence analysis of cryosectioned muscle from the index case, confirming the presence of fibers lacking complex I (NDUFB8) and complex IV (COXI) protein. Each dot represents the measurement from an individual muscle fiber, color coded according to its mitochondrial mass (blue-low, normal-beige, high-orange, and very high–red). Gray dashed lines indicate SD limits for the classification of fibers. Lines next to x- and y-axes represent the levels (SDs from the average of control fibers after normalization to porin/VDAC1 levels; _z= Z-score, see Methods section of Rocha et al. 2015 for full description of statistics7) of NDUFB8 and COX1, respectively (beige = normal [>−3], light beige = intermediate positive [−3 to−4.5], light purple = intermediate negative [−4.5 to −6], and purple = deficient [<−6]). Bold dotted lines indicate the mean expression level observed in respiratory normal fibers. (C) Western blot analysis of AAC and OXPHOS complex subunits on control and patient skeletal muscle samples. AAC = ADP/ATP carrier; ADP = adenosine diphosphate; ATP = adenosine triphosphate; COX = cytochrome c oxidase; GADPH = Glyceraldehyde 3-phosphate dehydrogenase; OXPHOS = oxidative phosphorylation; SDH = succinate dehydrogenase.