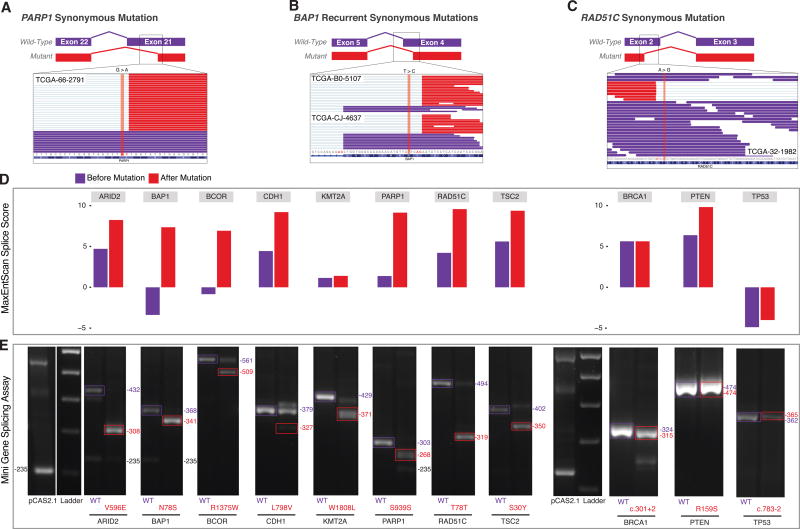
Figure 5. Minigene Functional Assay of Splice-Site-Creating Mutations.
(A) Integrative genomics viewer (IGV) screenshot of the conventionally annotated synonymous mutation in PARP1 in exon 21. RNA-seq reads of the candidate splice-site-creating mutation reveal the creation of an alternative splice site (red reads) created by the conventionally annotated synonymous mutation.
(B) Candidate recurrent splice-site-creating mutations in BAP1. Conventionally annotated as synonymous variants, the BAP1-mutated region shows alternatively spliced reads (red reads) in the IGV screenshot for each sample with the splice-site-creating mutation.
(C) IGV screenshot of a conventionally annotated synonymous mutation in RAD51C in exon 2.
(D) Maximum entropy score of the splice-site-creating variant before (purple) and after (red) the introduced mutation for each variant functionally validated in the mini-gene splicing assay. In silico predictions suggest all mutations strengthen the alternative splice site.
(E) Candidate splice-site-creating mutations validated by the mini-gene splicing assay. Exons of interest were cloned into the pCAS2.1 vector and mutant (red); wild-type (purple) plasmids were transfected into 293T cells; and total RNA was extracted to identify mutation-induced alternatively spliced products.