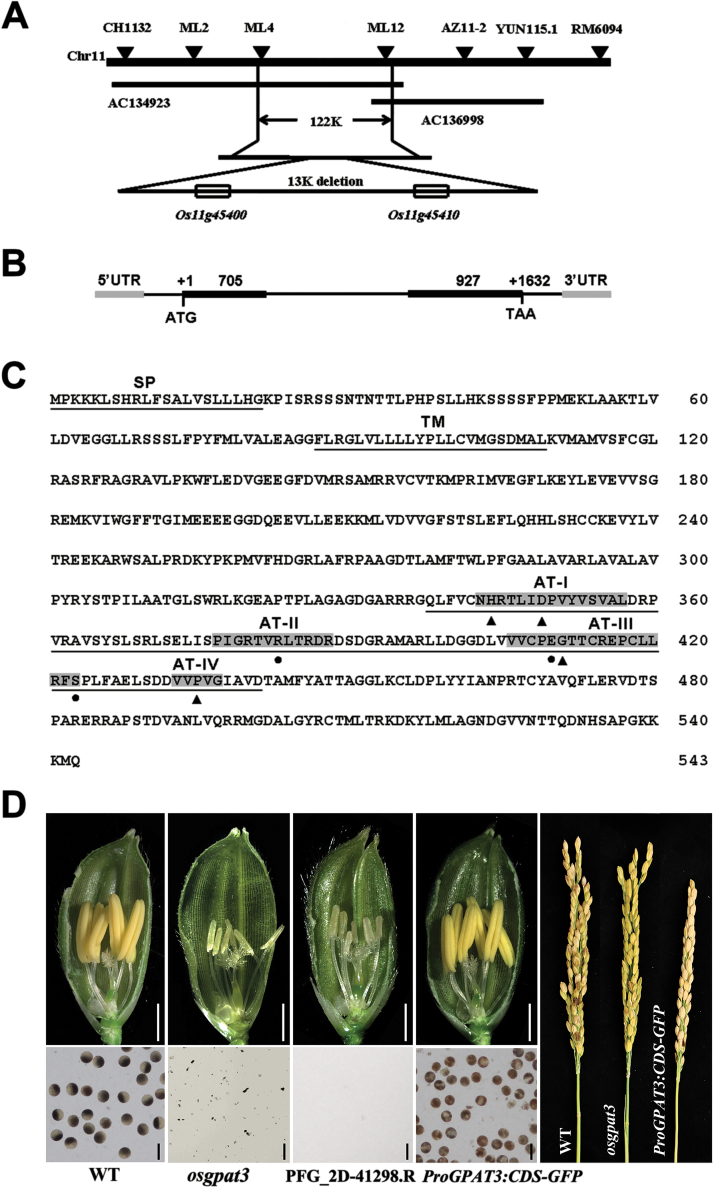
Fig. 6.
Molecular identification and sequence analysis of OsGPAT3. (A) Fine mapping of the osgpat3 mutation on chromosome 11. The names and positions of the molecular markers are indicated. AC134923 and AC136998 are genomic DNA accession numbers. The mutation was mapped to a 122 kb region between two molecular markers (ML4 and ML12). (B) A schematic representation of the exon and intron organization of LOC_Os11g45400. (C) Predicted protein sequence of OsGPAT3. The protein is predicted to contain a 21 amino acid signal peptide (SP), a transmembrane domain (TM), and a conserved acyltransferase domain (AT) with four conserved motifs. Dots indicate key amino acids important for substrate binding and triangles indicate key amino acids important for catalysis. (D) Functional complementation of osgpat3 with the full-length CDS of LOC_Os11g45400 fused to GFP, driven by its own promoter (3 kb). The LOC_Os11g45400 T-DNA insertion homozygous mutant PFG_2D-41298.R also shows a similar male sterility phenotype to that of the osgpat3 mutant. WT, wild type. Bars=2 mm for spikelets and 50 μm for pollen grains. (This figure is available in colour at JXB online.)