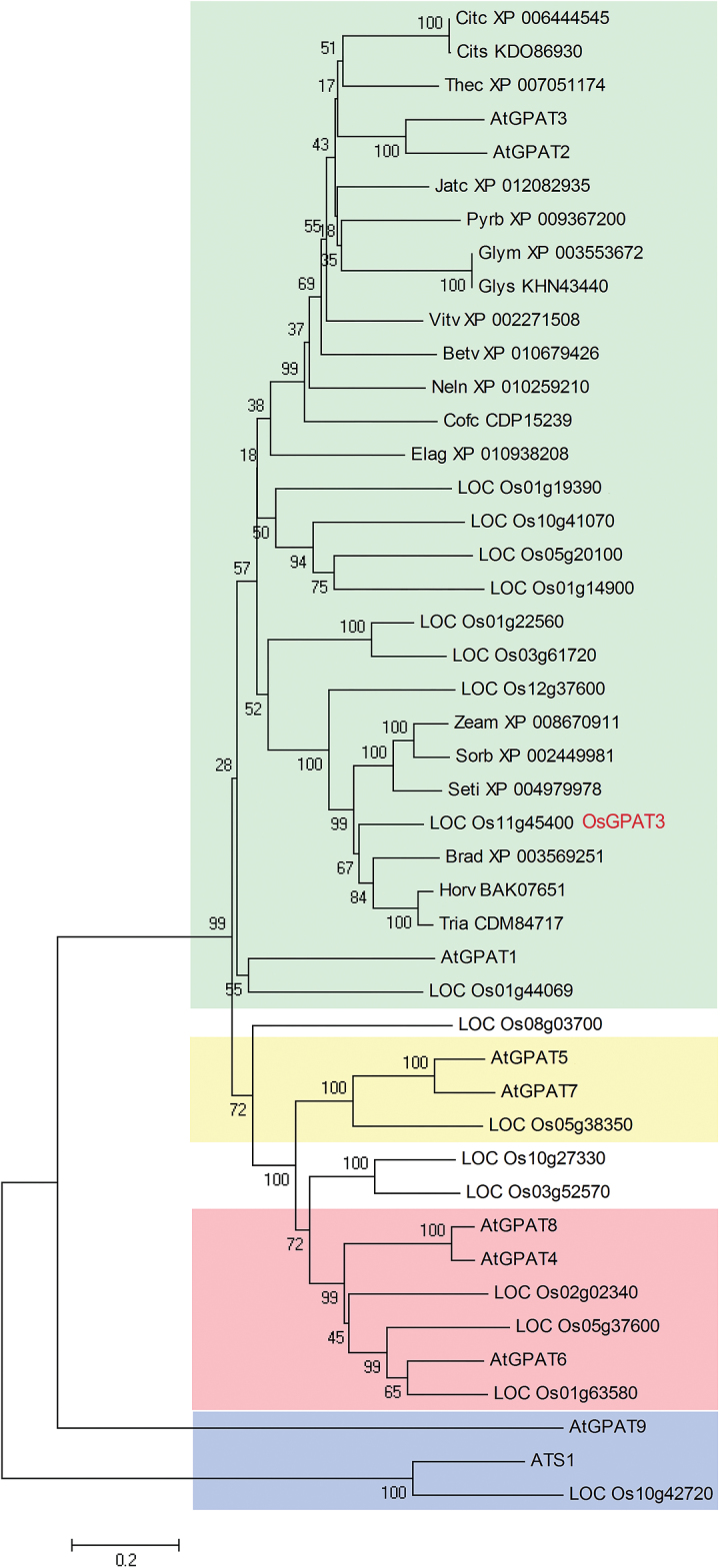
Fig. 7.
Phylogenetic tree of the GPAT family and OsGPAT3-related orthologs. A neighbor-joining phylogenetic tree was constructed with of all the Arabidopsis and rice GPATs and OsGPAT3 orthologs from other 18 species (E-value less than 7e-132) found in the NCBI, RGAP, and TAIR databases. The proteins are named according to their gene names or NCBI accession numbers. Bootstrap values are percentage of 1000 replicates. The length of the branches is proportional to the amino acid variation rates. Different clades are highlighted in different colors, representing the sn-1 GPAT (blue), the GPAT4/6/8 clade (red), the GPAT5/7 clade (yellow), and the GPAT1/2/3 clade (green). At, Arabidopsis thaliana; Os, Oryza sativa; Citc, Citrus clementina; Cits, Citrus sinensis; Thec, Theobroma cacao; Jatc, Jatropha curcas; Pyrb, Pyrus x bretschneideri; Vitv, Vitis vinifera; Glym, Glycine max; Glys, Glycine soja; Betv, Beta vulgaris; Neln, Nelumbo nucifera; Cofc, Coffea canephora; Elag, Elaeis guineensis; Zeam, Zea mays; Sorb, Sorghum bicolor; Seti, Setaria italica; Brad, Brachypodium distachyon; Horv, Hordeum vulgare; Tria, Triticum aestivum. (This figure is available in colour at JXB online.)