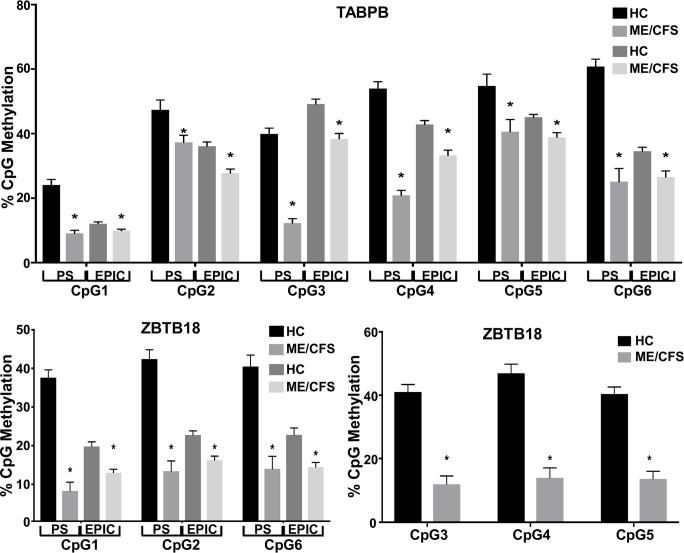
Fig 7. Bisulfite-conversion followed by pyrosequencing (PS) validation of DMPs identified by Illumina MethylationEPIC microarrays (EPIC).
Average methylation level of CpG sites within promoters of TABPB and ZBTB18 were assessed using pyrosequencing. Corresponding probes for TABPB: CpG1—cg04415168 (TSS1500), CpG2—cg10376053 (TSS1500), CpG3—cg14288848 (TSS1500), CpG4—cg17055704 (TSS1500), CpG5—cg14473643 (TSS1500), CpG6—cg14309283 (TSS1500). Corresponding probes for ZBTB18: CpG1—cg16399365 (TSS1500), CpG2—cg15896892 (TSS1500), CpG6—cg19698993 (TSS1500). CpG3, CpG4 and CpG5 are not printed on the Illumina MethylationEPIC microarrays and are located in ZBTB18 promoter between CpG2 and CpG6. * = FDR ≤ 0.05 for EPIC; * = p ≤ 0.05 for PS. Error bars represent the standard error of the mean.