Figure 2.
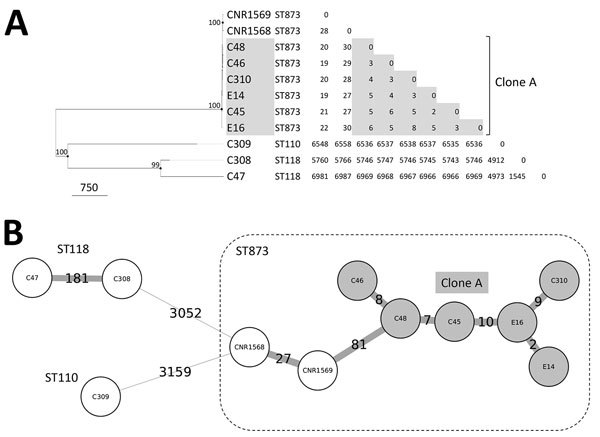
Whole-genome typing of Enterobacter cloacae complex isolates from nosocomial outbreak involving carbapenamase-producing Enterobacter strains, Lyon, France, January 12, 2014–December 31, 2015. A) Dendrogram inferred by the maximum-likelihood method on the basis of core genome SNPs. The node sizes are proportional to the bootstrap values; values >80 are indicated. Scale bar indicates SNPs. The relatedness of the strains was determined by using <15 variant sites as clonality criteria. B) Minimum-spanning tree based on a whole-genome multilocus sequence typing approach, combining the analysis of core genome loci and the presence or absence of accessory genes. Labels on branches indicate the absolute number of variant loci (clonality threshold <10 variant loci). SNP, single-nucleotide polymorphism; ST, sequence type.
