Figure 7.
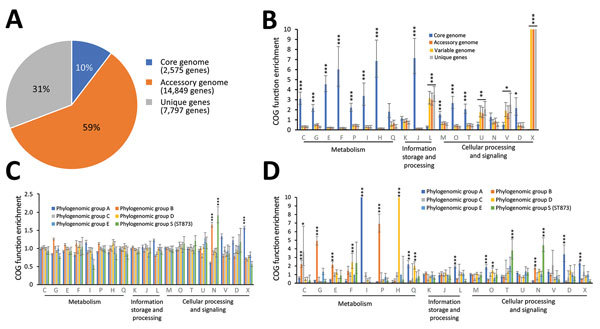
Pangenome analysis of metacluster Enterobacter hormaechei in study of nosocomial outbreak involving carbapenamase-producing Enterobacter strains, Lyon, France, January 12, 2014–December 31, 2015. A) Distribution of COGs); B) functional annotations in the pangenome; C) functional annotations in the variable genome (accessory genome + unique genes); and D) functional annotations for specific genes. Bar charts show the enrichment of COG categories as odds ratios; error bars indicate 95% CIs. Asterisks indicate certain COG categories that are significantly enriched: *p<0.05; **p<0.01;***p<0.001, all by Fisher exact test. Each COG category is identified by a 1-letter abbreviation: C, energy production and conversion; D, cell cycle control and mitosis; E, amino acid metabolism and transport; F, nucleotide metabolism and transport; G, carbohydrate metabolism and transport; H, coenzyme metabolism; I, lipid metabolism; J, translation; K, transcription; L, replication, recombination and repair; M, cell wall/membrane/envelope biogenesis; N, cell motility; O, post-translational modification, protein turnover, and chaperone functions; P, inorganic ion transport and metabolism; Q, secondary metabolism; T, signal transduction; U, intracellular trafficking and secretion; V, defense mechanisms; and X, mobilome. COG, clusters of orthologous groups.
