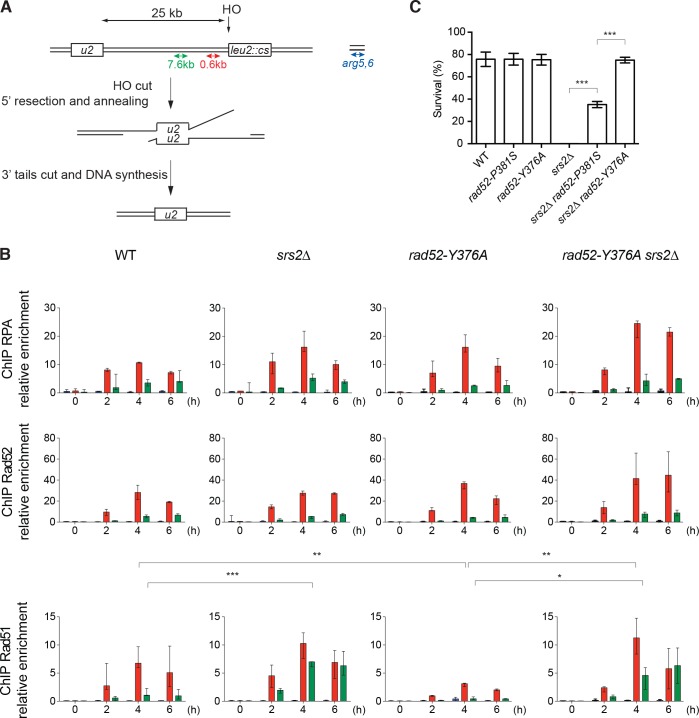
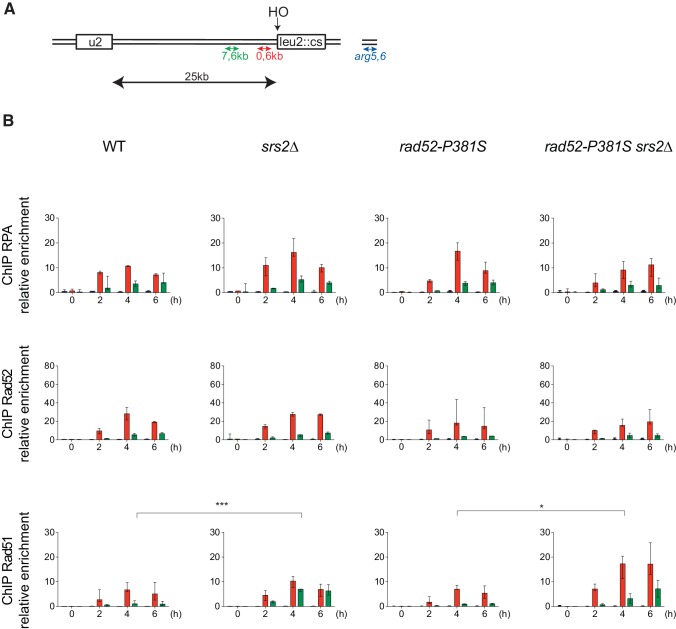
(A) Schematic representation of the HO-induced DSB repair system used. Breakage by HO can be repaired by single strand annealing (SSA) of two directed repeats located 25 kb apart. (B) ChIP analysis after HO induction in wild type (WT) or Srs2-deficient cells that express WT or Rad52-P381S-FLAG proteins. The median value of at least three experiments is shown and error bars represent the upper and lower values. Fisher’s exact test, *p<0.05, **p<0.01, ***p<0.001. Statistical analysis was performed at the 4 hr time point; only significant differences are shown. The rad52-P381S mutation did not significantly change Rad51 or Rad52 enrichment at 7.6 kb (green) and 0.6 kb (red) from the DSB, but a 1.5-fold significant increase in RPA recruitment was observed compared with WT Rad52. This increase could be related to an enrichment of ssDNA coated with both RPA and Rad51. This higher RPA enrichment is dependent on Srs2 presence because RPA loading is reduced by 1.8-fold in rad52-P381S srs2Δ cells compared with rad52-P381S cells. This is associated with a 2.5-fold increase in Rad51 loading at 0.6 kb from the DSB site in rad52-P381S srs2Δ cells compared with rad52-P381S cells. This suggests that Srs2 removes significantly more Rad51 in rad52-P381S cells. Similarly, Rad51 loading at the 7.6 kb site was increased by 3.4-fold in rad52-P381S srs2Δ cells compared with rad52-P381S cells. Therefore, Rad51 filaments assembled by Rad52-P381S are sensitive to Srs2 activity regardless of the distance from the break, confirming that Rad52 association with Rad51 protects Rad51 filaments from Srs2. Interestingly, the recruitment of Rad51 is increased specifically in rad52-P381S srs2∆ cells compared with rad52-P381S cells and srs2∆ cells for a reason we do not understand.