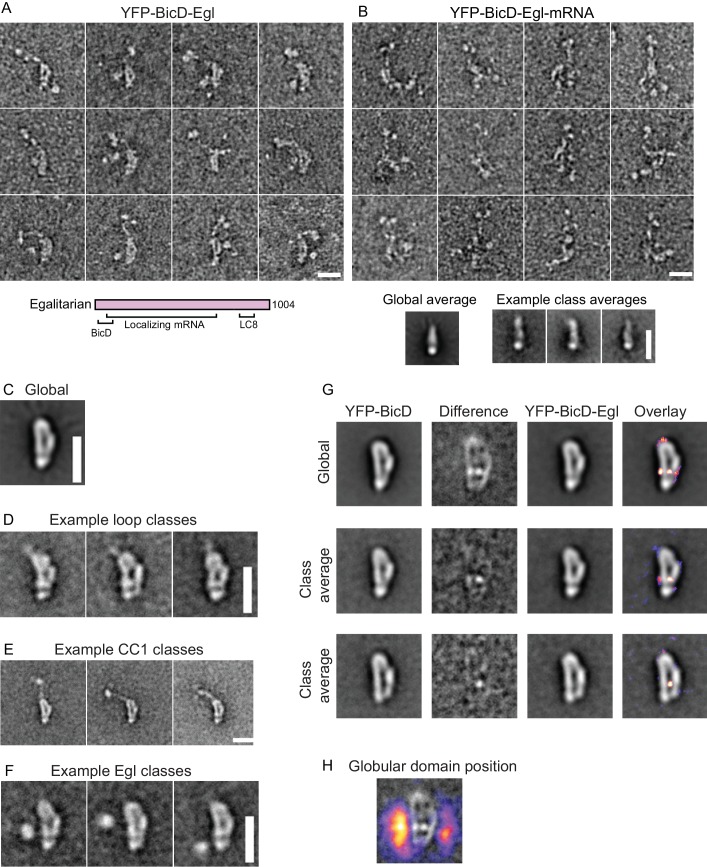
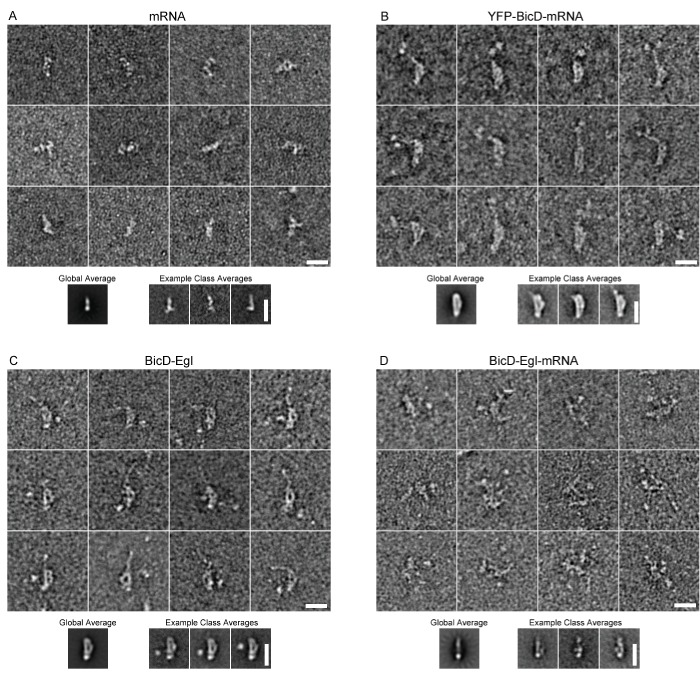
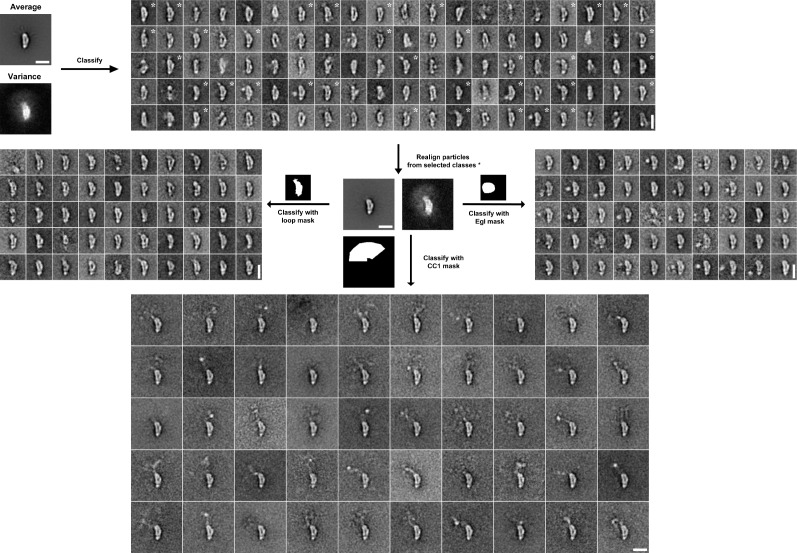
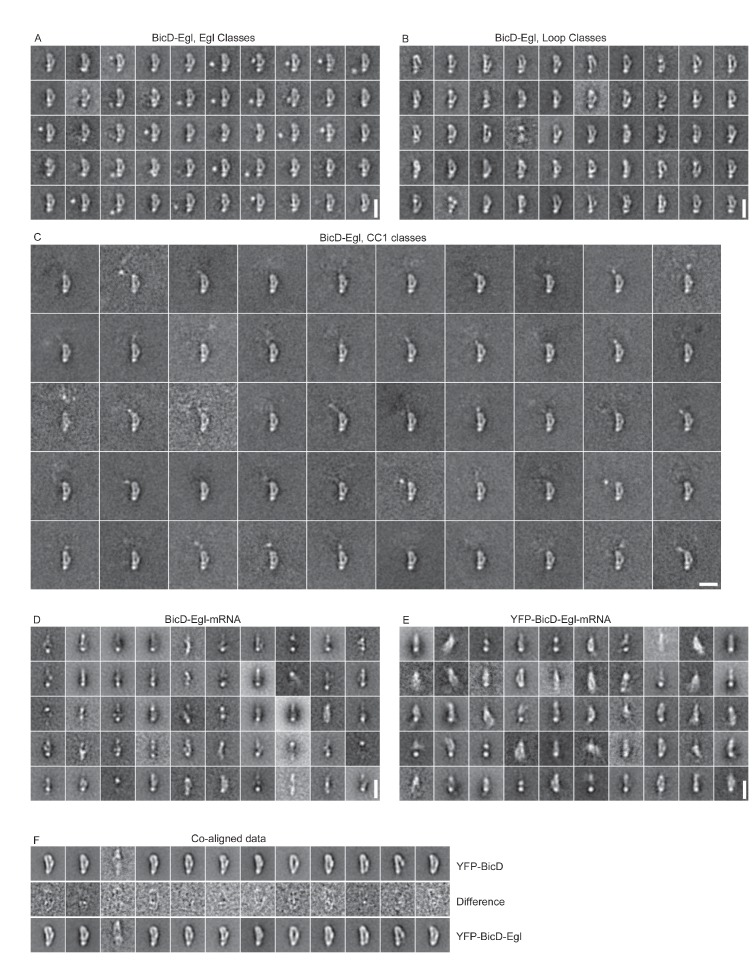
Figure 3. Electron micrographs of the YFP-BicD-Egl and YFP-BicD-Egl-mRNA complex.
(A) Montage of images showing that BicD retains the auto-inhibited looped conformation in the presence of bound Egl. A schematic of Egl (below) shows that the N-terminal domain binds BicD, the C-terminus binds dynein light chain (LC8), and mRNA binding is mediated through a large number (~800) of amino acids. (B) Montage of images showing that YFP-BicD no longer retains the auto-inhibited looped conformation in the presence of bound Egl and mRNA (K10min). Global and class averages are shown below the montage. (C) Average of all YFP-BicD-Egl molecules. (D) Example class averages of YFP-BicD-Egl with classification focused on the loop. (E) Example class averages of YFP-BicD-Egl with classification focused on the protruding YFP-CC1 region. (F) Example class averages of YFP-BicD-Egl with classification focused adjacent to the loop, to reveal the globular domain of Egl. (G) Co-alignment and classification of YFP-BicD and YFP-BicD-Egl. Upper row shows the average of YFP-BicD and YFP-BicD-Egl images (after an initial alignment, classification and selection of particles in similar orientations). Difference maps are shown in the second column and the difference map is overlaid as a heatmap onto the average of YFP-BicD-Egl in the fourth column, to show clearly the locations of greatest difference. Second and third rows show the averages and differences for individual classes. See Figure 3—figure supplement 5 for the full classification. (H) Heatmap showing the position of the globular Egl domain. Scale bars = 20 nm.
Figure 3—figure supplement 1. SDS-PAGE gels of Egl.