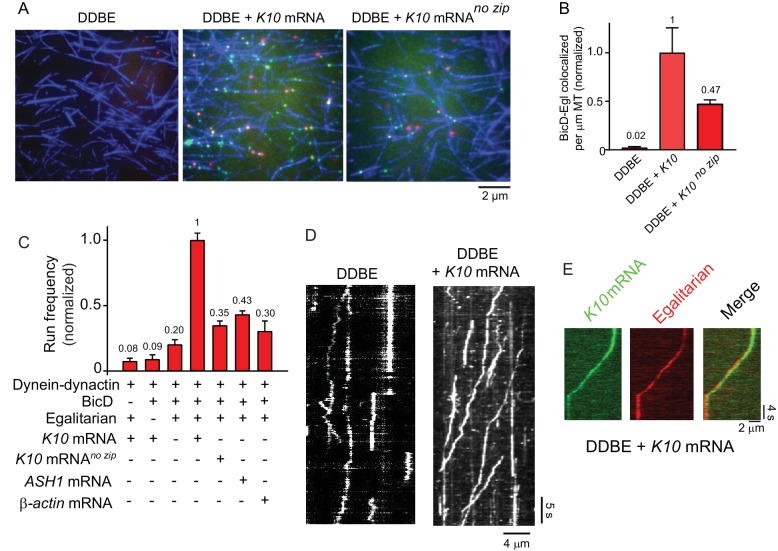
Figure 4. K10 mRNA is needed for BicD-Egl to recruit dynein-dynactin for motility.
(A) Single-molecule pulldowns of YFP-BicD (green) and Qdot-Egl (red) by dynein-dynactin (DDBE) bound to microtubules (blue) in the absence or presence of K10 mRNA. (B) Quantification of the single-molecule pulldown data showing the normalized number of colocalized BicD-Egl complexes bound to dynein-dynactin per µm microtubule length in the absence of mRNA (DDBE), in the presence of K10 mRNA (DDBE + K10), or K10 without the TLS zip code (DDBE + K10 no zip). (C) Normalized run frequencies (per μM dynein per μm microtubule length per time) of motile mRNPs. (Bars, left to right) Little movement of labeled K10 mRNA in the absence of either BicD or Egl. Few events are also observed for mRNPs lacking mRNA (movement was visualized with a Qdot bound to Egl). Fully reconstituted mRNPs with K10 mRNA have the highest run frequency, while mRNPs reconstituted with K10 mRNA lacking the TLS zip code showed reduced run frequencies. Two unrelated mRNA constructs, S. cerevisiae ASH1 mRNA and mammalian β-actin mRNA showed run frequencies similar to that of K10 mRNA lacking the TLS zip code. Error bars, sem, n ≥ 4 movies per condition, two independent experiments. (D) Kymographs illustrating motion of dynein-dynactin-BicD-Egl (DDBE) in the absence or presence of K10 mRNA. (E) Kymograph showing moving DDBE-K10 mRNA complexes dual labeled with Egl bound to a Qdot (red) and K10 mRNA labeled with Alexa Fluor 488-UTP (green). See Figure 4—source data 1.